BioByte 147: A New Approach for Biological Foundation Models, Microbubble Robots Navigate Tumor Environments, and RESPLICE Rewrites RNA
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
Biology needs to become prospective [Arcadia Science, The Stacks, February 2026]
Why it matters: Biological Foundation Models (BFMs) are experiencing a pseudoreplication crisis due to data imbalances that give rise to underlying biases. Brute force scaling by adding new sample data does not fully address this issue; in all likelihood, a different solution is needed: an informed stance regarding where new data needs to be collected. Researchers at Arcadia suggest a prospective, Bayesian approach to data collection which allows BFMs to participate more actively in the scientific process, allowing experimentalists to work to close the gaps in biological knowledge via this more directed approach.
The early 2000s saw the rise of biology’s first big data era with the advent of significant cost reductions and throughput increases for genomic sequencing. This first explosion of data abundance led to the emergence and subsequent growth of data-oriented disciplines within the greater field of biology, such as bioinformatics, computational biology, systems biology, and all fields of -omics. Emerging as the pinnacle of years of work from these fields, Biological Foundation Models (BFMs), have become drivers of massive biological data collection due to their increased capability to synthesize large datasets and draw novel insights. Some particularly staunch supporters have even speculated that, “The internal representations learned by BFMs may reflect deep principles about the organization of life.”
While immensely powerful, researchers at Arcadia purport that the meteoric growth of BFMs may signal the end of this first era of big data for biology. Their argument takes the stance that much of the data used to train these models is rife with fundamental distribution imbalances which result in apparent increases in performance without actually correcting any of the underlying biases. This effect often arises from overrepresented sequences, which if uncorrected, can result in pseudoreplication. The predominant answer to this threat and its associated consequence has so far been to increase the size of the training data, although usually this occurs without regard for the composition of the added data itself.
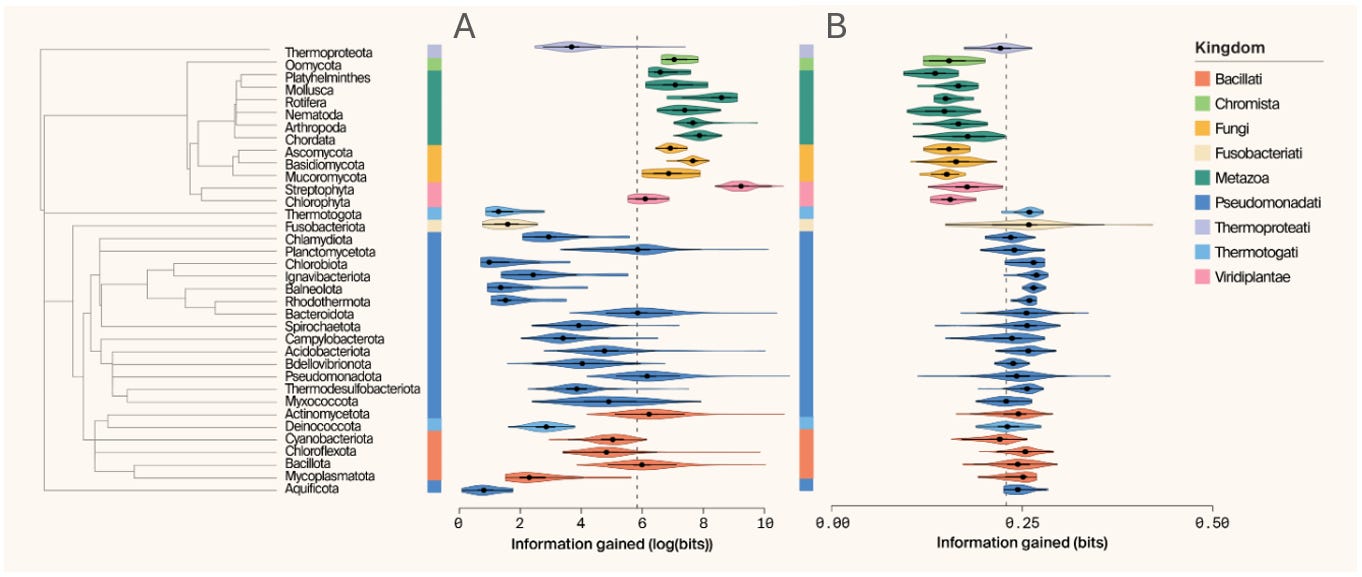
As an alternative, Arcadia emphasizes the importance of using an informed, prospective approach to data accumulation and model training via a Bayesian framework. This method mandates being explicit regarding (i) the prior knowledge, (ii) how new observations update that knowledge, and (iii) the specific quantity to be optimized. These points help us answer the pivotal directedness required for their Bayesian framework. They illustrate how this works via the AlphaFold Database (AFDB). As it stands, AFDB is dominated by a small number of bacterial taxa which generally yield higher-confidence (pLDDT) predictions while other lineages—particularly within the eukaryotes—are much more sparsely sampled. The important point they note is that this is not ameliorated by simple reweighting to balance data; it must be solved by elucidating where to collect new data. This suggests the need for an initial decision to be made regarding unit analysis (e.g. individual proteins, Foldseek clusters, environmental samples, or whole proteomes), termed the estimand, which is here defined as “the information provided by a new proteome”. Derived from Kullback-Leibler (KL) divergence between the prior and posterior Dirichlet distributions, the quantity thus represents the information gain (IG).
To exemplify this, scientists at Arcadia first examined IG by taxa, ignoring proteome size. In this analysis, eukaryotic phyla are overarchingly represented with a mean IG of 4999 bits versus bacterial (523 bits) and archaeal (166 bits) phyla. However, if you alter this analysis to breakdown by random samples in fixed-sized subsets (n=100) from each species instead of by whole proteomes, this pattern is reversed with prokaryotic phyla now exhibiting higher IG than eukaryotes. This demonstrates the potential usefulness of broad microbial sampling for exploration of the protein structural space. Overall, the conclusion here echoes the purpose of the sampling: if looking to optimize for prediction across species where efficiency is paramount, microbial data is the best option for this breadth; on the other hand, if looking to maximize for structural novelty, eukaryotic proteomes—given their larger quantities—is best for this depth.
”The end of biology’s first big data era isn’t a signal to stop measuring; rather, it’s a mandate to start measuring differently.”
Employing a prospective framework ultimately enables greater utility of the data, suggesting a means to circumvent the current diminishing returns experienced by BFMs. In this approach, researchers calculate the expected information gain before a sample is sequenced, allowing for better efficiency and cost-effectiveness of data collection while also reintroducing BFMs as “active participants in the scientific process” as opposed to existing as passive consumers of large-scale, opportunistic data accumulation. Precision and informed data collection will drive the next generation of biological discovery, allowing biologists—aided by BFMs—to truly discover what Arcadia puts as “the rare, the divergent, and the truly novel.”
Papers
Enzymatic microbubble robots [Tang et al., Nature Nanotechnology, February 2026]
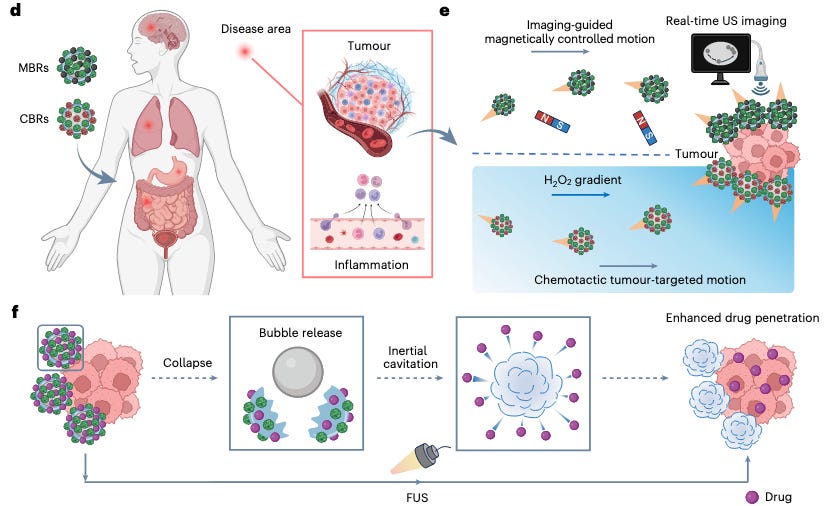
Why it matters: Tang et al. describe a new platform to develop microbubble robots that can autonomously navigate to tumor environments using magnetic nanoparticles and biocatalysis. Combined with ultrasound-trigged delivery mechanisms, this technology offers a powerful solution to critical barriers in targeted drug delivery efforts to treat a host of diseases in challenging tissue environments.
While all the rage these days seems to center around drug discovery, another significant challenge is the problem of drug delivery, especially in tissues like the bladder, brain, and GI tract. To that end, micro- and nanoscale approaches have emerged as promising contenders, with the field seeing a “shift from traditional metallic or inorganic robots to bioinspired integrations” using a range of biological components like mammalian cells and membrane vesicles to ensure better biocompatibility, degradation, and overall control. In this joint paper from the Gao and Shapiro labs at Caltech, the authors describe a “cost-effecive, multifunctional microbubble robot platform” that improves upon previous complex fabrication processes. The authors use this platform to develop two types of microbubble robots that use biocatalysts as an autonomous propulsion system and use their internal microbubble to serve as an ultrasound imaging contrast agent and drug delivery mechanism. These MBRs were then evaluated in bladder cancer tumor models where they showed “substantially improved antitumor efficacy.”
The team demonstrated the versatility of their platform by developing two types of urease-powered robots: magnetically controlled bubble robots (MBRs) and chemotactic bubble robots (CBRs). Using bovine serum albumen (BSA) microbubbles as a base, magnetic nanoparticles (MNPs) were incorporated onto the bubble surface using relatively simple electrostatic adsorption, followed by the addition of modified urease enzymes to the BSA shell and MNPs using click chemistry. When placed in an environment like the bladder, the urease enzymes convert urea into ammonia and carbon dioxide to facilitate self-diffusiophoretic (movement by creating its own local concentration gradient) propulsion. Furthermore, the MNPs allowed for “magnetic guidance enabled steering” compared to otherwise random movement. Using similar design methods, the team also designed CBRs to investigate the potential of autonomous motion control using chemical sensing. CBRs were functionalized with catalase alongside the urease enzymes to sense and navigate toward naturally occurring hydrogen peroxide gradients at tumor sites. Importantly, both MBRs and CBRs utilize their internal gas cores as contrast agents for real-time tracking and can be triggered to deliver therapeutic payloads by ultrasound-induced cavitation.
After verifying the survivability and response of MBRs and CBRs to different control methods, the authors then verified that the robots could actually carry a drug payload without losing structural integrity and motility. Using the chemotherapeutic agent doxorubicin (DOX) for testing, the team used electrostatic adsorption to have the drug stick to the bubble surface rather than inducing chemical bonding. This method achieved a loading efficiency of just over 30% with there being no reduction when urease and catalase enzymes were added to the bubble exteriors. In vitro tests on T24 bladder cancer cells showed that CBRs showed superior adherence to cancer cells compared to bare bubbles, demonstrating that the addition of self-propulsion systems helped achieve physical contact and sustained proximity to target cells. The team also ensured that the focused ultrasound pulses required to induce cavitation and delivery were within FDA approved safety limits; furthermore, the collapsed protein shells showed good biodegradation, being eliminated completely within twelve hours when exposed to trypsin. Finally, the bubble robots were tested in cancer spheroids and proper live mice models. Tests showed that robots treated with focused ultrasound were able to achieve deeper penetration into spheroids and orthotopic bladder tumors in mice. Finally in mice models, the team tested the effects of passive bubbles (no additional motion modalities), MBRs, and CBRs when combined with focused ultrasound (FUS). Tests showed that passive bubbles with DOX and FUS treatment were not able to achieve significant tumor reduction, while active robots without FUS were able to show greater effect. Combining CBRs and FUS showed the most effective reduction, proving that the combination of FUS and autonomous movement was crucial to the platform’s results.
Rewriting endogenous human transcripts with dual CRISPR-guided 3′ trans-splicing [Chandrasekaran et al., Cell Systems, February 2026]
Why it matters: Chandrasekaran et al. introduce RESPLICE, a programmable RNA rewriting technology that enables targeted mRNA editing through exon replacement. Instead of permanently editing DNA, the system transiently rewrites endogenous mRNA using RNA-targeted CRISPR effectors, which reduces risks associated with genomic damage while enabling correction of large or heterogeneous mutations that are difficult to address with base or prime editing. The platform can splice in replacement exons up to ~2 kb and achieves therapeutically relevant editing efficiencies in endogenous human transcripts.
DNA editing is powerful but permanent and can introduce cellular damage or off-target genomic changes. RNA editing is transient, tunable, and potentially safer because it avoids altering the genome. However, most current RNA approaches rely on antisense oligos or knockdown strategies that mainly modulate splicing or expression rather than replacing large coding regions. The authors instead develop programmable trans-splicing, where engineered RNA cargo is stitched into endogenous pre-mRNA (think of it as targeted pre-mRNA replacement, but extending all the way to the end of the molecule). Their system uses catalytically dead Cas13d (dCasRx) to shuttle a trans-splicing donor RNA to a specific transcript and a second RNA-targeting nuclease to deplete competing endogenous cis-splicing. Because this replaces entire transcript segments, it can correct multiple mutations simultaneously across large regions that are difficult to address with current DNA gene editing technologies.
The team first built a dual-fluorescent reporter to quantify on-target versus off-target trans-splicing. If the correct pre-mRNA is replaced, cells show green fluorescence; if the wrong pre-mRNA is replaced, they show blue fluorescence. This first setup reached ~30% trans-splicing efficiency (fraction of target transcripts edited) with ~95% on-target specificity. They then hypothesized that removing competing cis-splicing products could further improve efficiency. They added a cis-splicing interfering module (CIM), which uses an active RNA-targeting CRISPR effector to cleave the native transcript and bias splicing toward the engineered donor, increasing efficiency roughly 1.2–2× depending on configuration. When targeting the endogenous ITGB1 gene, adding the CIM improved trans-splicing efficiency from ~0.2–3% to ~40%, and up to ~90% in cells expressing high levels of the editing components.
For therapeutic proof of concept, they targeted hereditary hemochromatosis, an iron overload disease caused by mutations in HFE that is currently treated only via lifelong phlebotomy. In Huh7 cells carrying an in-frame Tyr271 deletion, they delivered replacement HFE exons and achieved ~8% trans-splicing. For context, a prior in vivo base editing study showed therapeutic benefit at ~10.7% correction of the pathogenic C282Y mutation. Here, trans-splicing replaces an entire exon region, which is not currently possible with most genetic editing tools, suggesting a path toward correcting broader mutation classes with a single therapy design.
Notable deals
Phylo launches from stealth with $13.5M seed to build AI-powered scientific assistants. The round was co-led by a16z and Menlo Ventures (via its Anthropic-partnered Anthology Fund). Founded by Stanford PhDs Kexin Huang and Yuanhao Qu, the startup is commercializing Biomni, an AI agent for biomedical research the pair developed during their doctoral work, handling tasks like guide RNA design by asking clarifying questions, proposing research plans, and returning ranked recommendations. Phylo joins a growing AI scientist space alongside Edison Scientific and Lila Sciences, competing for enterprise pharma and biotech deals against frontier lab offerings like Anthropic’s Claude for Life Sciences.
Colossal Biosciences secures nine-figure UAE partnership to launch world’s first global genetic BioVault network. The deal includes a $60M equity investment – extending Colossal’s Series C and bringing total capital raised to $615M – plus revenue tied to the company’s cryogenic preservation platform. The facility will use cryogenic storage, robotics, and AI-powered monitoring to bank living cell lines, tissue samples, and genomic data from 10,000+ species, initially targeting the 100 most imperiled animals not currently biobanked elsewhere. An open data initiative will make non-proprietary genomic information available to researchers worldwide.
The Department of Energy launches OPAL, a multi-lab initiative to build autonomous AI-driven biofoundries as part of the Genesis Mission, bringing together Berkeley Lab, Oak Ridge, Argonne, and Pacific Northwest national laboratories with industry partners Teselagen and FutureHouse. The project uses robotic systems, AI agents, and standardized data-sharing platforms to accelerate biotechnology, targeting applications in biomanufacturing, agriculture, and critical mineral recovery. OPAL’s initial focus is building general-purpose biology foundation models for microbial engineering that link genes to organism function, then integrating these models with automated lab infrastructure to run experiments autonomously that would otherwise take weeks or years.
Illumina completes $350M acquisition of SomaLogic to deepen proteomics capabilities and advance multiomics strategy. The acquisition excludes Standard BioTools’ mass cytometry and microfluidics businesses, with Illumina taking only the aptamer-based and functional proteomics assets including KREX and Single SOMAmer technologies. The combined portfolio integrates SomaScan with Illumina Protein Prep, DRAGEN software, and Illumina Connected Multiomics to generate multiomic datasets at scale. For Standard BioTools, the sale simplifies operating structure and moves the company toward break-even on an adjusted EBITDA basis.
Automata raises $45M Series C to scale AI-ready lab automation. The round was led by Dimension with participation from Danaher Ventures, Tru Arrow Partners, Octopus Ventures, and Entrepreneurs First. Automata now counts five top pharma companies as customers. Funds will scale deployments, build next-gen closed-loop experimentation software, and expand global operations. Beckman Coulter Life Sciences and Molecular Devices will integrate their instruments into Automata’s ecosystem.
Cellares closes $257M Series D to scale automated cell therapy manufacturing globally. The round was co-led by BlackRock and Eclipse, with new investors T. Rowe Price, Baillie Gifford, Duquesne Family Office, Intuitive Ventures, EDBI, and Gates Frontier joining existing backers. The company’s Cell Shuttle platform, the first to receive FDA Advanced Manufacturing Technology designation, delivers end-to-end, closed-system cell therapy production with ~10x higher throughput and lower per-patient costs than conventional CDMOs. Clinical manufacturing begins H1 2026; commercial-scale production in 2027. IPO targeted for Q4 2027.
Loyal raises a $100M Series C led by age1 to advance lead canine longevity asset in clinical trials. The asset, LOY-002, takes the form of a daily prescription pill that proactively targets the underlying metabolic mechanisms behind aging. Already in the past year it has already completed two of the three major requirements for the drug’s application for Expanded Conditional Approval (XCA): the Reasonable Expectation of Effectiveness (RXE) and Target Animal Safety (TAS) sections as well as the STAY study, a pivotal trial and the largest yet in veterinary medicine, enrolling 1,300 dogs across 70 clinics in the US. Should approval be granted, LOY-002 would not just be the first FDA-approved drug for lifespan extension in canines but the first in any species. This move is predicted to have tremendous impact for the longevity space and paves a way for broader longevity-targeted therapeutics in humans. Funding will be used to prepare for market launch as well as to further build out the Loyal team and distribution channels. Baillie Gifford and existing investors also participated in the round.
Takeda enters into a multi-year partnership with Iambic Therapeutics around AI drug discovery in a new deal worth up to $1.7B in milestone payments.In the deal, Takeda plans to leverage Iambic’s AI drug discovery and development engine to further certain high-priority small molecule targets. Under focus initially are oncology, gastrointestinal, and inflammation-related indications although this may very well expand given the duration of the partnership. Additionally, Takeda will gain access to NeuralPLexer, Iambic’s proprietary protein-ligand complex predictive ML model as well as the company’s fully-integrated high-throughput automated wet lab platform, allowing for rapid ‘Design-Make-Test-Analyze’ cycles. Per the deal terms, Iambic will receive undisclosed amounts in upfront, research cost, and technology access payments as well as the up to 1.7B in success-based milestone payments and net sales royalties on any assets produced in this collaboration.
In case you missed it
The Isomorphic Labs Drug Design Engine unlocks a new frontier beyond AlphaFold
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.