BioByte 146: How Pleiades Predicts Alzheimer's, New EDEN Models Enable Drug Design via Evolutionary Datasets, Exploring the Inverse Relationship between Peripheral Cancer and AD, and Synthetic Ecology
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
Using Interpretability to Identify a Novel Class of Alzheimer’s Biomarkers [Wang et al., Goodfire, January 2026]
Why it matters: Researchers from Goodfire AI utilize mechanistic interpretability methods to uncover the features that Pleiades, an epigenetics foundation model, uses to successfully predict Alzheimer’s Disease (AD) from cell-free DNA (cfDNA) samples.
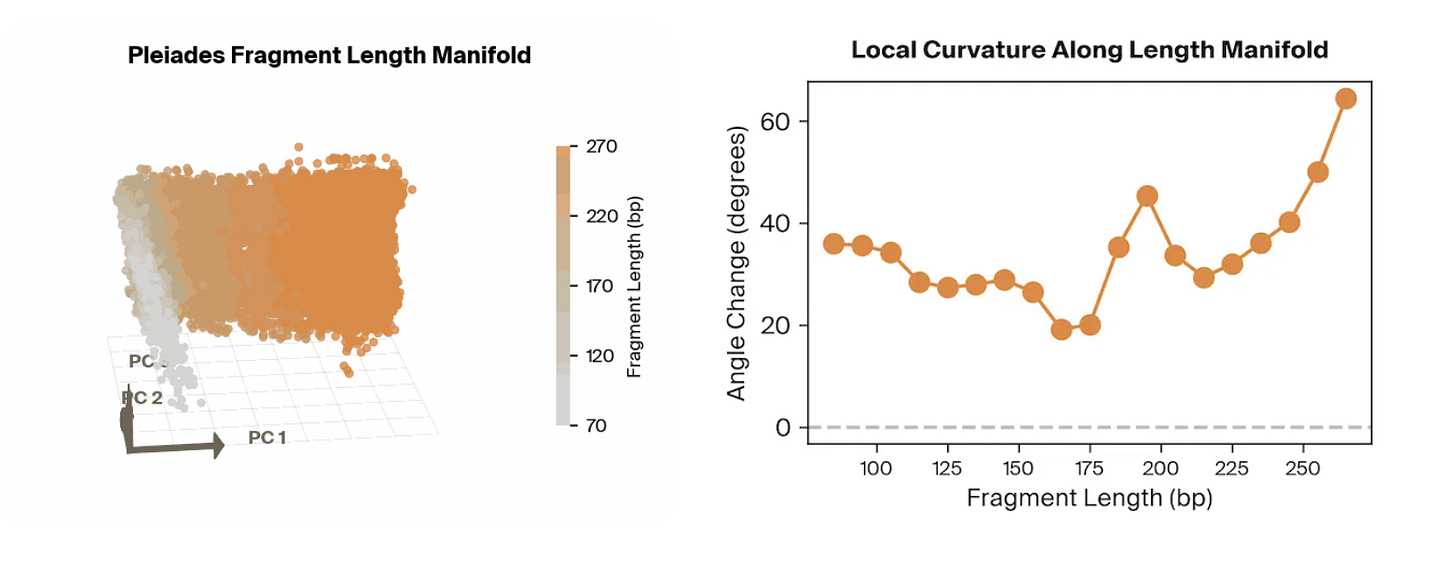
Pleiades is an epigenomic foundation model developed by Prima Mente that captures methylated and unmethylated human DNA from full genomes and cfDNA. They utilize a hierarchical set-attention architecture that enables them to expand attention across a large swath of DNA, such as for pools of cfDNA. In the original preprint, one powerful application Pleiades was pointed towards was the prediction of whether or not a patient had AD based on their cfDNA sequences, which it did extremely well in with an AUROC of 0.82.
The team at Goodfire applied mechanistic interpretability techniques to the frozen, cfDNA-fine-tuned Pleiades model, probing its final sample-level representations to identify what features enabled it to effectively predict AD status. Rather than relying on post-hoc feature attribution on raw inputs, they decomposed the internal embedding space using linear probes and sparse autoencoders to recover what qualities the embeddings contained.
Surprisingly, the strongest features utilized by the model were all correlated to the length of the input cfDNA sequences. With a simple classifier that takes in fragment length features alone, they achieved an AUROC of approximately 0.78 on an independent cohort. By combining fragment length with other biologically meaningful signals identified through their probing, such as methylation- and cell-type–related features, they achieved performance around 0.84 AUROC, approaching the full Pleiades cfDNA-only performance of ~0.82–0.83. More broadly, this work demonstrates how mechanistic interpretability can turn opaque foundation-model representations into concrete, testable biological hypotheses, helping bridge the gap between high-performing black-box predictors and mechanistic understanding of the disease signals they exploit.
Papers
Peripheral cancer attenuates amyloid pathology in Alzheimer’s disease via cystatin-c activation of TREM2 [Li et al., Cell, January 2026]
Why it matters: Alzheimer’s disease therapies have largely focused on limiting the formation of amyloid plaques. However, once plaques are established, the brain has limited capacity to remove them, and most interventions show diminishing returns in symptomatic disease. At the same time, epidemiological studies have long noted an inverse correlation between cancer and Alzheimer’s incidence, though this has generally been attributed to confounding factors. This work provides a concrete mechanistic explanation linking peripheral cancer to active amyloid clearance in the brain.
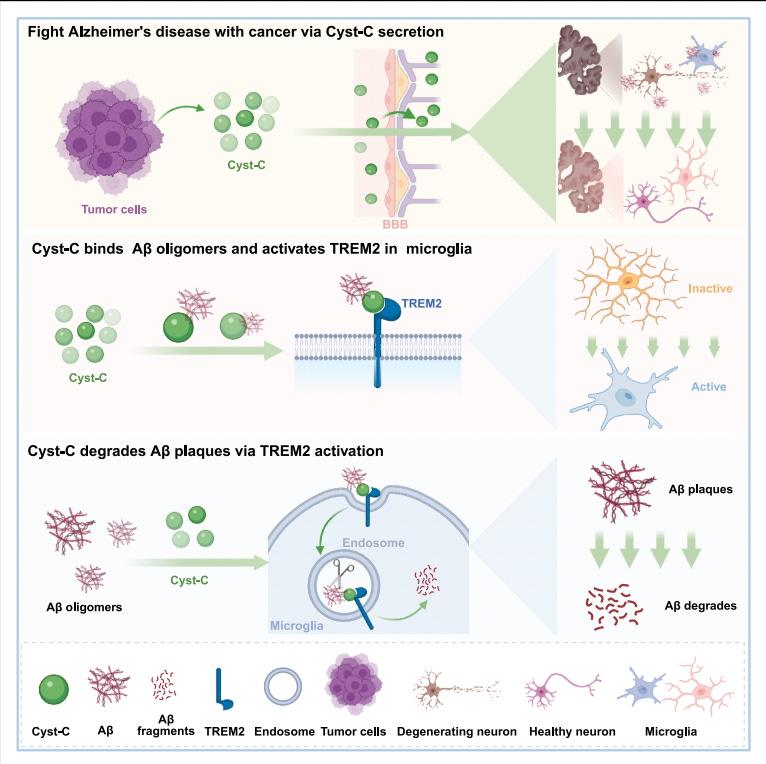
Li et al. show that multiple peripheral tumors secrete cystatin-C (Cyst-C), a small circulating protein that crosses the compromised blood–brain barrier in Alzheimer’s models and accumulates in amyloid plaques. In mouse models of Alzheimer’s disease, the presence of lung, prostate, or colon tumors substantially reduces plaque burden and rescues cognitive performance. Importantly, injecting only the tumor secretome reproduces the same effect, indicating that the phenomenon is mediated by circulating factors rather than tumor physiology itself.
Through proteomic profiling and genetic perturbation, the authors identify Cyst-C as the necessary and sufficient effector. Deleting Cyst-C from cancer cells abolishes amyloid clearance, while reintroducing human Cyst-C restores it. Exogenous administration of recombinant Cyst-C is sufficient to drive plaque reduction even in aged mice with established pathology.
Mechanistically, Cyst-C binds amyloid oligomers and engages the microglial receptor TREM2, activating the DAP12–SYK signaling axis and shifting microglia into a phagocytic, lysosome-active state. In vivo, Cyst-C localizes to plaques, increases the fraction of activated CD68⁺ plaque-associated microglia, reduces TMEM119⁺ resting microglia, and accelerates degradation of pre-existing deposits. The effect is strictly dependent on this pathway: plaque clearance is lost in TREM2 knockout mice, in mice carrying the AD-associated TREM2 R47H mutation, or when using a disease-associated Cyst-C mutant (L68Q) that fails to bind TREM2. Functionally, this establishes a clearance mechanism that operates downstream of amyloid deposition. Rather than modifying amyloid production or aggregation kinetics, Cyst-C couples existing aggregates to the brain’s innate immune machinery and enables their active removal.
This work expands the therapeutic design space for neurodegeneration. It suggests that established protein aggregates can be treated as removable substrates through targeted engagement of endogenous clearance pathways, and that TREM2-directed adapter-like molecules may provide a general strategy for reversing pathological accumulation.
Designing AI-programmable therapeutics with the EDEN family of foundation models [Munsamy et al., bioRxiv, January 2026]
Why it matters: The EDEN-family of foundation models demonstrates the potential of combining massive evolutionary datasets with powerful language modeling architectures for improved performance on a range of therapeutically relevant design challenges.
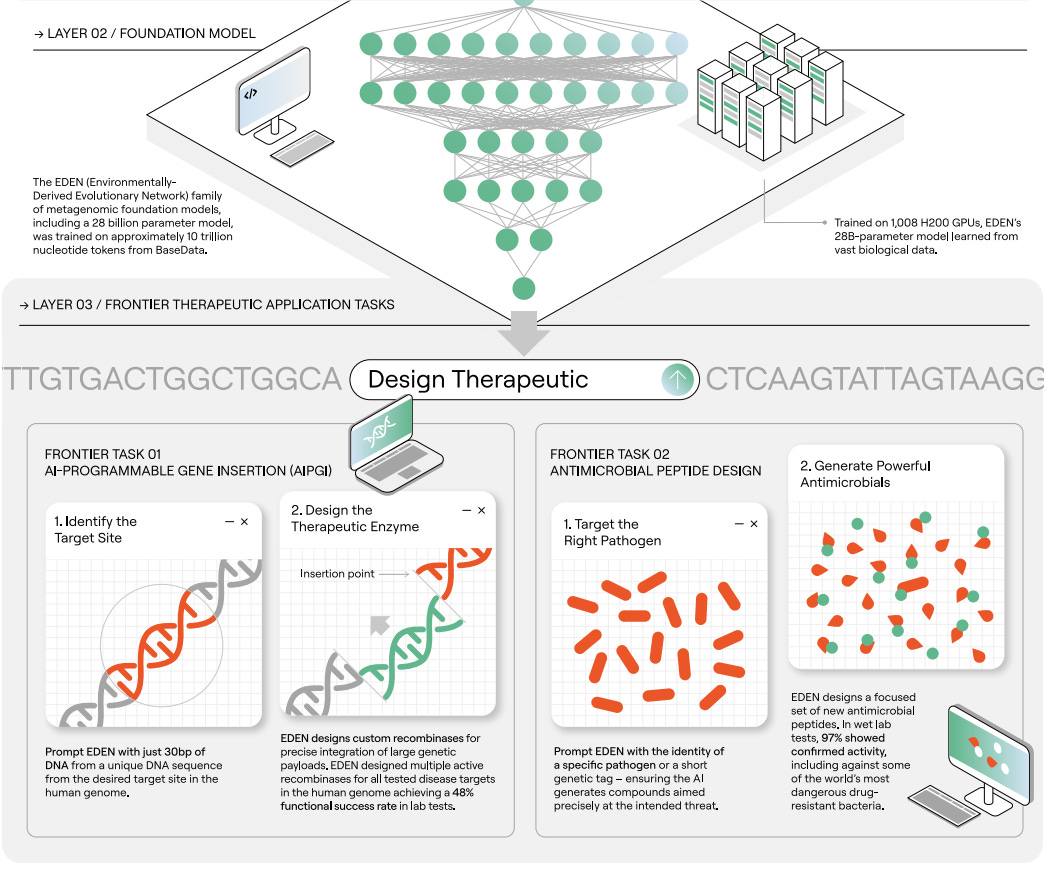
Earlier this month, Basecamp Research presented the EDEN (environmentally-derived evolutionary network) family of foundation models, aimed to address the “profound disparity between information content within biological systems and the processing bandwidth of…engineering tools.” Operating under the premise that the “inability of current models to reliably execute complex tasks necessary for multi-modality therapeutic design may arise from a fundamental deficit in the scale and diversity of the available training data,” the company first curated the BaseData dataset meant to capture the genomes of over 1 million species absent in other databases, with nearly 10 billion novel genes. Building on this massive data corpus, the authors developed the EDEN family, aiming to tackle a range of challenges across generative genomics and protein design.
Built using a Llama3.1-style architecture, EDEN models were trained on BaseData using a next-token autoregressive objective. To compare the impact of BaseData on downstream model quality, the team also trained versions of EDEN on OpenGenome-2, the data used for training Evo 2. Tests showed that smaller models trained on OG2 showed superior performance while larger models were better suited to BaseData. The authors hypothesized this trend was driven by small models “[underfitting] the longer-range structure in the longer metagenomic assemblies in BaseData” while larger models could instead exploit. Prior to conducting more novel design challenges, the authors benchmarked EDEN’s zero shot performance on mutational effect prediction and a suite of gene completion tasks, with the models showing state of the art performance on the former and comparable results to Evo 2 on the latter with notably unique sequence space coverage.
The most interesting demonstration of EDEN’s capabilities came from testing its potential therapeutic utility, with the authors targeting large gene insertion, antibiotic peptide design, and microbiome design. The team first set out to design large serine recombinases (LSRs) and bridge recombinases (BRs) for large gene insertion tasks. Prompting the models with 30 nucleotides representing the desired target site, the authors generated multiple active LSRs for a variety of disease-associated genomic loci. The team selected twenty LSR designs and found that over half were capable of insertion into therapeutically relevant T cells, with the best design showing activity comparable to wild type proteins. Interestingly, EDEN was also able to design active BRs when prompted with non-coding guide RNA sequences, with some candidates showing only 65% sequence similarity to those in training data. Next, the team focused on antimicrobial peptide (AMP) design by prompting the model with pathogen-specific tags that could nudge the model towards certain targets. Notably, 97% of tested AMPs showed some activity, with the top candidates having low micromolar potency against multidrug-resistant pathogens. Finally, EDEN was tasked with generating a fully synthetic microbiome. The authors fine-tuned EDEN with a single digestive system microbiome sample from BaseData and then generated 100,000 sequences (10kb in length) to yield a gigabase sized synthetic microbiome. This output was then characterized at the sequence level and taxonomic level, with 99% of the identified taxonomic units being consistent with known other species found in the human gut. Pathway enrichment analysis showed that the synthetic microbiome could mimic crucial processes involved in digestion. Interestingly, EDEN also generated sequences consistent with viral prophages known to be present in microbiome bacteria, demonstrating that the model had learned patterns of host-phage relationships. In summary, this trio of tasks was a positive first step towards proving the therapeutic potential of EDEN-like models across a range of design tasks.
Statistical design of a synthetic microbiome that suppresses diverse gut pathogens [Oliveira et al., bioRxiv, December 2025]
Why it matters: Designing live microbial therapeutics to displace pathogens in the gut is a critical issue for infectious disease, post-antibiotic recovery, and engineered microbiome medicine. However, the combinatorial space of possible consortia is astronomical and mechanistic priors (e.g. microbe-microbe interactions, metabolic networks) are often incomplete – failing to scale to the complexity of real communities. Oliveira et al. provide a statistical design framework that infers functional community structure directly from experimental screens. They apply it to build a statistically distilled synthetic microbiome – SynCom15 – that suppresses multi-drug resistant Klebsiella pneumoniae as effectively as whole-fecal transplants in pre-clinical models. This lays the groundwork for experimentally-efficient, data-driven design of emergent microbial functions – e.g. synthetic ecology.
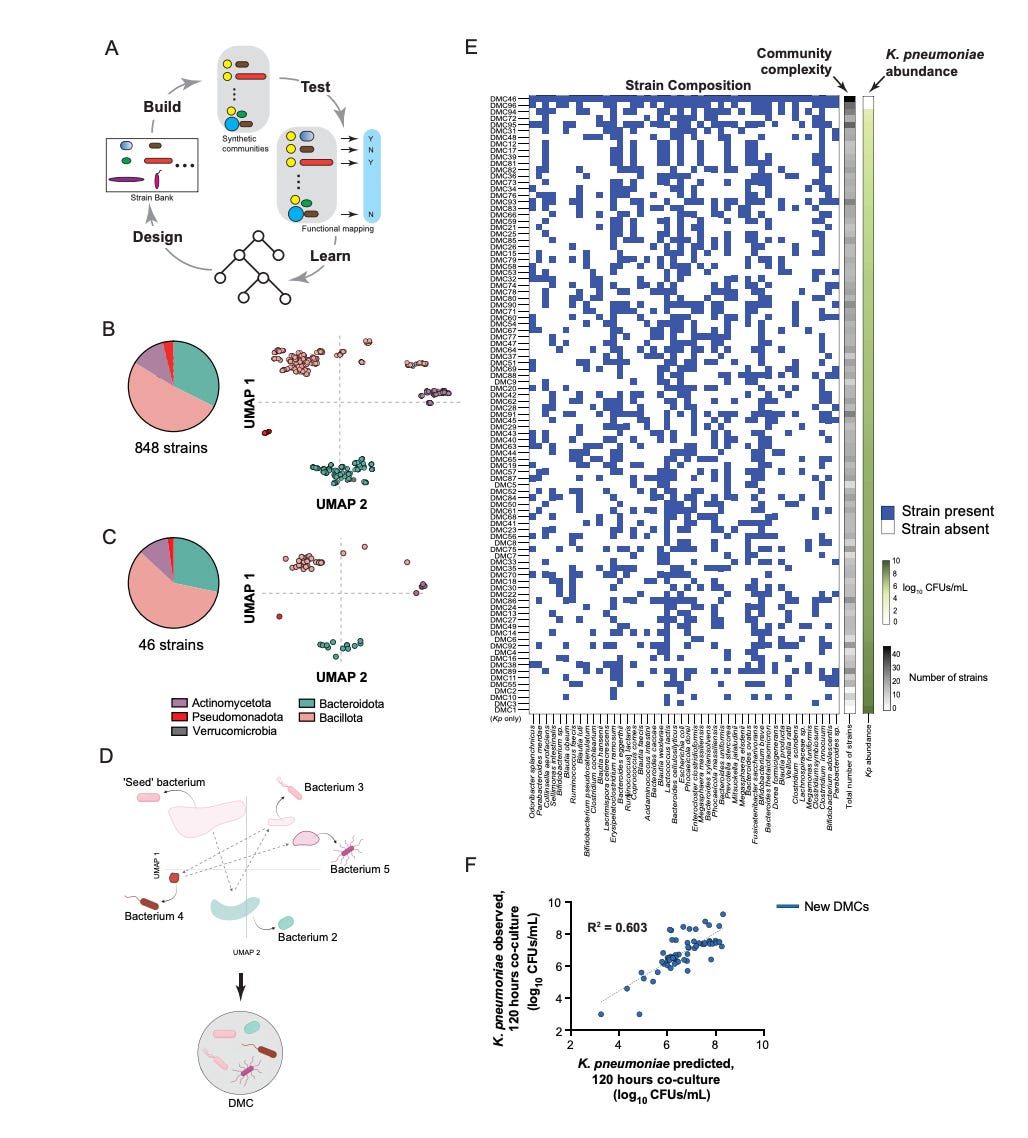
The team compressed a bank of 848 human gut commensal strains into 46 representative strains and assembled 96 diverse metagenomic consortia (DMCs) by systematically sampling combinations through a dimensionality reduction representation (UMAP) of genomic diversity. Rather than typical iterative cycles, they ran a single screen where the 96 DMCs were each co-cultured with the pathogen. This enabled them to implement ‘constraint distillation’, a framework where binary presence / absence patterns of strains – not detailed mechanistic features – served as the input for spectral analysis via singular value decomposition to identify low-dimensional constraints predictive of pathogen suppression. The model distilled this space into a generative rule set that identified a 15-strain core highly predictive of suppression.
The consortium formed by these strains – SynCom15 – yielded clearance of K. pneumoniae across diverse environments – BHIS media, germ-free cecal media, and antibiotic-treated murine cecal extracts, reducing pathogen abundance to below the detection limit by day seven of treatment. Mechanistically, this suppression was driven by fatty acid production coupled with environmental acidification, a contact-independent method that also cleared other pathogens like C. difficile. In contrast, models based on community metabolic profiles performed poorly – overfitting to sample-specific artifacts.
Beyond this case, the approach hints at a generalizable route to synthetic ecology: using high-throughput experimental sampling to generate a labeled dataset of pathogen suppression and learn conditional, constraint-driven generative models that predict emergent community function without detailed priors – akin to ML-guided design seen across materials and drug discovery.
With validation of robustness across host genotypes and diets and evaluation of this constraint-distillation approach on other complex targets (e.g. metabolic modulation or immune engagement), this pipeline may meaningfully accelerate development of rational therapeutics for antibiotic resistance and other microbiome-related conditions.
Notable deals
Mendra launches with an oversubscribed $82M Series A co-led by Orbimed, 8VC, and 5AM Ventures. The San Francisco-based biotech is seeking to utilize AI to accelerate the development and commercialization of therapeutics for rare diseases. Founded by ex-BioMarin, Palantir, Modis Therapeutics, and Escient executives, Mendra is pairing deep sector knowledge of the rare disease space with AI-driven capabilities to tackle what founder, Joshua Grass, asserts are some of the biggest challenges of bringing rare disease drugs to market: asset selection, clinical development, and global commercialization. To this end, the company’s AI initiatives will be directed toward patient identification and clinical trial enrollment, as well as access to global markets. Funding from this round is declared to target acquisition and development of rare disease assets in building out Mendra’s initial portfolio. Other participating investors in the Series A include Lux Capital and Wing VC.
TRex Bio announces close of an oversubscribed $50M funding round to advance clinical Treg portfolio. Trex is targeting autoimmune and inflammatory diseases via regulatory T cells (Tregs)—in studying Treg behavior in human tissue, the aim is to uncover and address the disease in the respective tissue where it presents, quieting the inflammatory process and restoring tissue homeostasis, according to their website. The company’s Deep Biology platform has previously led to collaborations with Eli Lilly and J&J for immunology disorders target-identification and development. Currently Trex has one wholly-owned asset, TRB-061, a selective TNFR2 agonist, in the midst of Phase 1a/1b clinical trials for atopic dermatitis. This fundraise is intended to help progress TRB-061 further through trials as well as prepare two other assets, TRB-071 and TRB-081, for commencement of Phase I clinical trials expected to take place in 2027. Specific indications for which these latter two assets are aimed at targeting have not yet been released. Participating investors in the round include new investors, Janus Henderson Investors, Balyasny Asset Management L.P. and Affinity Asset Advisors, as well as existing investors Alexandria Venture Investments, Avego BioScience Capital, Delos Capital, Eli Lilly and Company, Johnson & Johnson Innovation, Pfizer Ventures, Polaris Partners and SV Health Investors.
Qilu Pharmaceuticals partners with Insilico Medicine in pursuit of cardiometabolic treatments in a new $120M deal.Through the partnership, the China-based pharma is hoping to leverage Insilico’s leading proprietary AI drug discovery platform, Pharma.ai, for joint development of small molecule inhibitors intended to manage cardiometabolic diseases. This notably follows Insilico’s new release during BIO-Europe of their expansive cardiometabolic portfolio which includes eight assets spanning hit to IND-enabling stages. Outlined in the deal are the respective roles of each party: while Insilico will focus on novel small molecule design and optimization, Qilu will handle subsequent development and commercialization. The $120M deal value includes milestone payments and single-digit royalties and marks the deepening of a collaboration between the two companies that initially began in 2021 when Qilu in-licensed Insilico’s PandaOmics, credited as a key contributor to the successful kickoff of the present partnership.
Primmune Therapeutics announces an $8.6M Series B extension to be used to fund Phase II trials for their lead asset. The compound, PRTX007, is a small molecule agonist of toll-like receptor 7 (TLR7), built to address solid tumors via moderation of the body’s immune response. It directs signaling towards a productive IRF7-driven poly-interferon response while suppressing pro-inflammatory cytokine pathways which can limit the efficacy of other drugs. Phase I studies saw the administration of the drug to 119 patients alongside an anti-PD-1 therapy, resulting in no serious adverse events and confirmation of desired immune signaling modulation. The Phase II trials will evaluate the efficacy of PRTX007 in Stage III resectable melanoma, still in combination with the anti-PD-1 therapy. Bioqube Ventures, Oberland Capital, and Samsara Biocapital participated in the round, which brings the Series B to a $23.3M total.
In case you missed it
The 1-in-a-Million Breakthrough Behind Genyro’s Vision for a Post-Darwinian Age
AI biotech, Hologen, founded by ex-Google CEO Eric Schmidt is raising $150 million
~180 lines of code to win the in silico portion of the Adaptyv Nipah binding competition
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.