BioByte 145: STACK Tackles Perturbation Effects, Magnetic Resonance for Biological Imaging, Revolutionizing DNA Assembly with Sidewinder, and Novel Therapeutic Strategies for Schizophrenia
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Papers
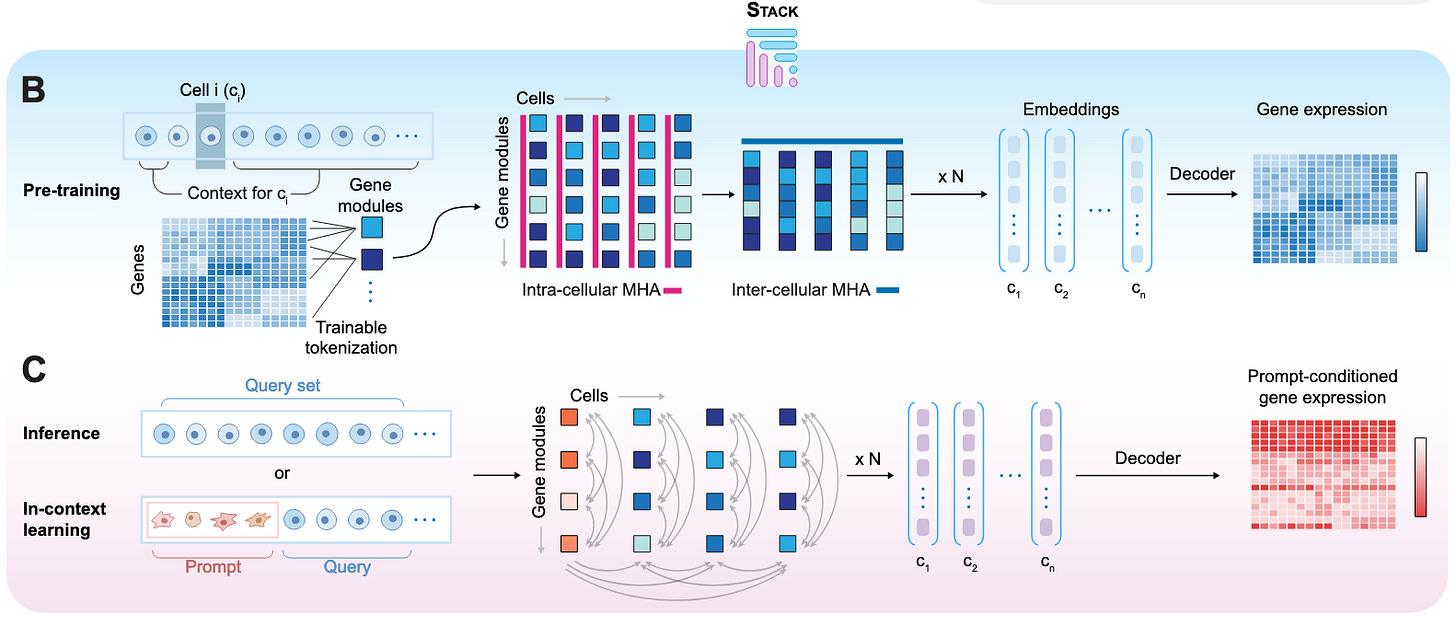
Stack: In-Context Learning of Single-Cell Biology [Dong et al., bioRxiv, January 2026]
Why it matters: STACK shows greatly improved zero-shot performance on various single-cell and perturbation effect modeling without the need for expensive finetuning. Its in-context learning pipeline is used to generate an atlas of perturbation effects across the human body that will hopefully serve as a useful in silico research resource.
Last year saw the continuation of large-scale dataset generation efforts in the realm of single-cell transcriptomics with a key focus on mapping perturbation effects. In 2025, companies like Tahoe Bio and Xaira Therapeutics released giant single-cell perturbation atlases like Tahoe-100M and X-Atlas/Orion, aiming to measure drug response effects and genome-wide dose-dependent perturbations respectively. Datasets like these and others from academia and industry alike are set to provide the high-resolution training data necessary to build the next generation of single-cell foundation models. However, even while currently available foundation models have grown to enormous sizes and shown that their representations are useful for downstream tasks, they still struggle to show meaningful improvement over classical approaches for zero-shot perturbation prediction baselines. Furthermore, such models require massive training datasets and supervised finetuning, limiting their practical use for work in unseen cell types and conditions. This is attributed to design choices in both training and architecture, where most models are trained explicitly at the single-cell level and can’t synthesize information across cell populations.
To tackle some of these challenges, Dong et al. present STACK, “a self-supervised framework that enables in-context learning through a novel architecture on cell sets.” At the heart of STACK lies a series of tabular transformer blocks that allows the model to leverage information both within a single cell and across an input cell population. STACK takes as input, a “cell set,” where every other cell with respect to some reference cell is treated as a cell’s context. The training pipeline then masks a randomly sampled set of genes across all cells and aims to predict gene expression across cells. A key component of the STACK framework is its “trainable tokenization at the gene-group level,” where instead of treating each gene in a cell as a distinct unit, the model compresses the latent space into 100 learned “gene module tokens.” This implicit grouping of genes into functional units is meant to force the model to retain crucial biological information and also helped with scaling to larger model sizes compared to a simpler gene-level tokenization strategy. The sum of these architecture choices enables greater control of the model’s predictions, which the authors provide as a reason for improved zero-shot performance and also leverage to incorporate in-context learning. Specifically, users can specify a desired biological state with “prompt” cells to predict the gene expression of a “query” cell population, allowing them to control for the predictions in query cells by modulating the prompt set and also predict the effects of perturbations.
STACK was put through an extensive testing regimen, with the authors first evaluating the quality of the model’s embeddings learned from cellular context at inference. When evaluated on observation samples and large-scale perturbation datasets of various modalities, STACK embeddings were found to mostly surpass alternative methods in predicting metadata like disease states and cell types when passed through linear and multi-layer perceptron probes. Next, the team assessed STACK’s capabilities on various novel in-context learning tasks such as perturbation effect in novel samples and novel cell types using the prompt/query pipeline. They also evaluated the model on observational tasks like held-out cell type prediction and cross-dataset cell type generation. STACK showed impressive performance, showing the best results on over 90% of the 31 benchmark tasks even against models like STATE (also from the same group at the Arc Institute in 2025) that were trained on orders of magnitude more supervised training data. Finally, the authors used STACK to “generate a perturbational whole-organism atlas (Perturb Sapiens)” through in-context learning. Specifically, they used a set of 90 cytokine perturbations and over 100 drug perturbations as prompts and the entire Tabula Sapiens human cell atlas spanning 28 tissues and 40 cell types as a query. This test was meant to measure some of the many cell-type-tissue-perturbation profiles that have never been acquired experimentally. The team used an internal MLP classifier to filter for high confidence predictions and used UMAP plots and log-fold-change metrics to identify cell-type specific and tissue-specific perturbation effects. Given the practical challenges of experimentally mapping perturbations across various cells and tissues, the authors hope that Perturb Sapiens can serve as an important resource for the in-silico study of drug effects across the body.
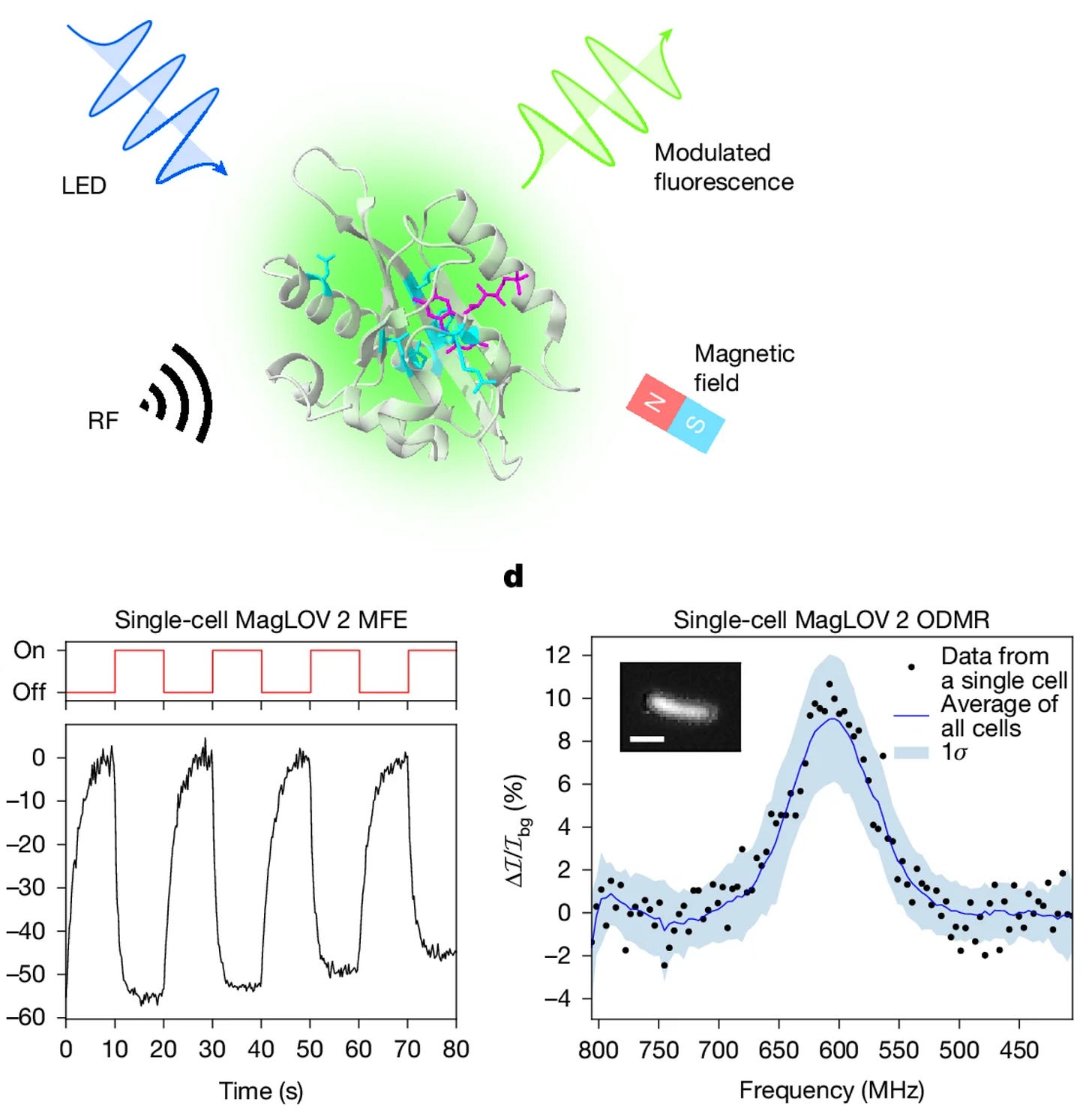
Quantum spin resonance in engineered proteins for multimodal sensing [Abrahams et al., Nature, January 2026]
Why it matters: The authors engineer a new class of genetically encoded reporters: magneto-sensitive fluorescent proteins (MFPs), whose fluorescence is modulated by electron-spin magnetic resonance. This introduces magnetic resonance as a completely new axis for biological imaging and sensing, opening the door to genetically encoded magnetic-field sensors, RF-detectable molecular barcodes, and MRI-like spatial encoding using magnetic field gradients.
Previously, scientists in Andrew York’s group (one of the authors) had shown that several flavin-binding fluorescent proteins weakly reacted to magnetic fields, slightly modifying their fluorescence. In that work, and continued in this work, they evolved those fluorescent proteins such that the strength of their response to magnetic fields grew much stronger. From a biotechnology perspective, this is intriguing because current genetically encoded sensors rely on light or chemistry. Optogenetics has transformed neuroscience, but is limited by scattering and absorption in tissue. On the other hand, magnetic fields and radiofrequency (RF) waves penetrate deeply and uniformly through tissue, and are already used to image deep tissue. If magnetic sensitivity could be genetically encoded and made strong, specific, and controllable, it would enable an entirely new modality of non-invasive sensing and control: magnetogenetics.
The team used directed evolution to turn weakly magneto-sensitive flavin proteins into robust quantum spin sensors. Starting from a LOV-domain protein scaffold, they iteratively mutated and screened variants under controlled magnetic and RF fields, selecting for proteins whose fluorescence strongly changed as their underlying electron-spin states were perturbed. As the result of two of their optimizations, they engineered a LOV-variant with a strong response to the magnet (MagLOV2) and one LOV-variant with a fast response to the magnet (MagLOV2-fast). These can be identified through their unique Hamiltonian “fingerprint” of their ODMR spectra (resonance frequencies, linewidths, and hyperfine structure).
To demonstrate the technology’s utility, they tested three settings where the new MagLOV proteins could be used as unique tags or biosensors. In one example, they used it as an orthogonal labeling channel in single-cell imaging. They mixed cells expressing conventional EGFP with cells expressing MagLOV (attached to mCherry so they could be identified). They alternated a static magnetic field on and off, measured the GFP-channel fluorescence over time, and extracted each cell’s magnetic-field dependent GFP modulation curve. MagLOV has a higher magnetic-field induced 450nm intensity variation than EGFP, and so these magnetic response curves were informative enough to correctly classify the MagLOV cells vs EGFP cells with ~99% accuracy, despite the fact that all of the cells shine green. In other experiments, they further demonstrated RF-driven ODMR in living cells and showed that applying magnetic field gradients shifts the resonance frequencies in a position-dependent manner (and therefore can be used as a spatial encoding).
The work is being spun into magnetically controlled therapies as the foundation of Nonfiction Labs. They’ve developed MagBodies, antibodies whose binding strength can be modulated by magnetic fields, potentially enabling magnetically inducible therapeutic binding events with spatial controllability.
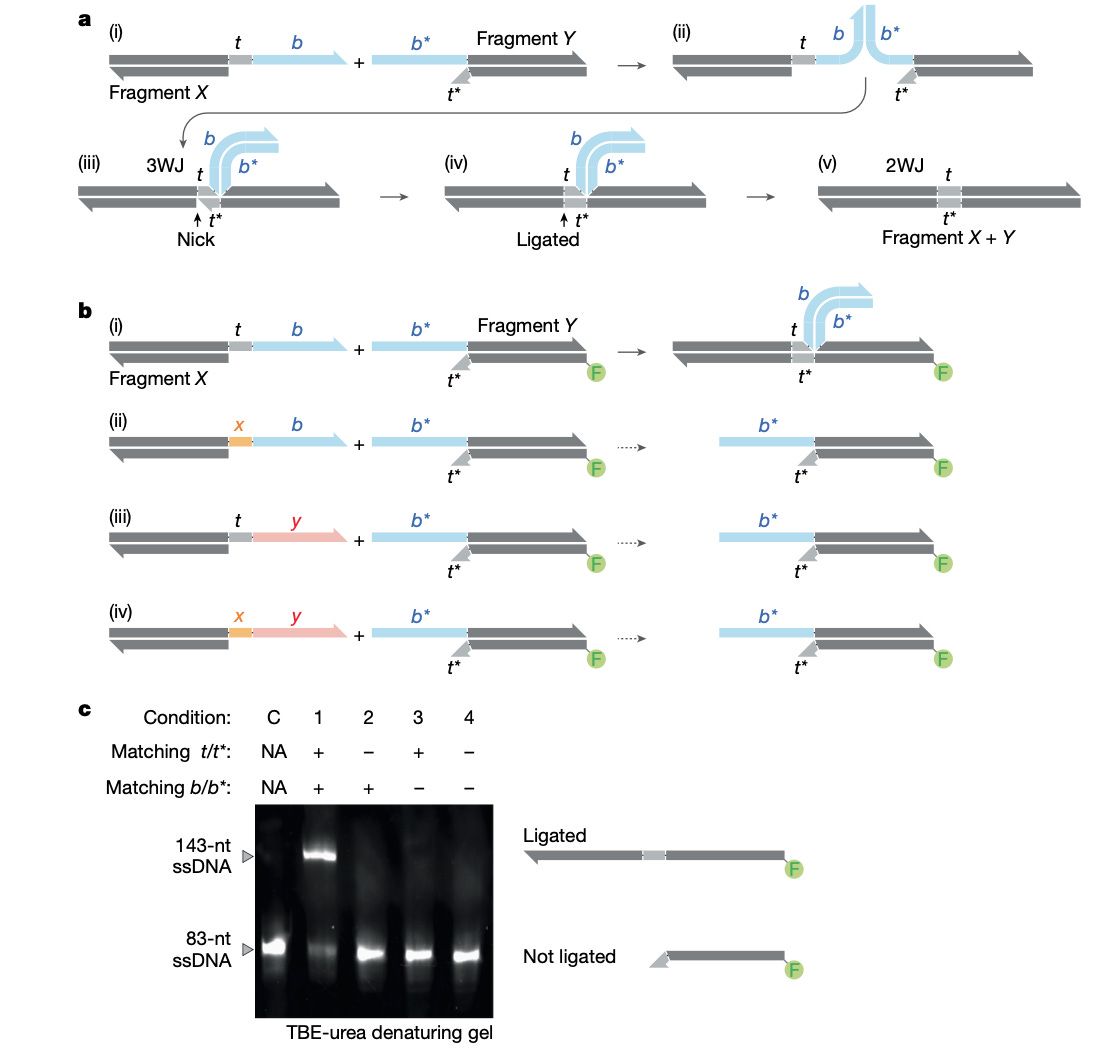
Construction of complex and diverse DNA sequences using DNA three-way junctions [Robinson et al., Nature, January 2026]
Why it matters: Biology can now be read and edited at an increasingly extraordinary scale, but building DNA – especially long, complex, or highly diverse sequences – bottlenecks synthetic biology. While emerging synthesis methods can directly write long DNA molecules, this scales additively – sequences are specifically constructed one at a time. In the face of combinatorial tasks (e.g. variant libraries), a multiplicative approach is needed: fragment assembly and reuse of shared DNA segments offer this scalable efficiency. However, there is a core architectural limitation: nearly all assembly methods (Gibson, Golden Gate, PCA) use sequence overlaps that become a part of the final construct, meaning the ‘instructions’ for the assembly are entangled with the sequence you’re aiming to build. This creates unavoidable constraints and failure modes when assembling many fragments, GC-heavy genes, repetitive elements, or large combinatorial libraries.
This paper introduces Sidewinder, a new DNA assembly paradigm that cleanly separates assembly guidance from the final DNA sequence using a DNA three-way junction (3WJ): fragments that carry short ‘toeholds’ that become a part of the product, but the high-specificity pairing that actually enforces correct ordering comes from longer barcode sequences that are not retained in the final construct. In other words, Sidewinder externalizes the ‘routing logic’ of assembly, enabling true ‘sequence-independent’ construction. Unlike sequence overlaps – which must be redesigned and validated for every new target and are constrained by the DNA being built – the same barcode scaffold can be used across arbitrary sequences without regard for sequence complexity, making assembly logic reusable.
Robinson et al. show that Sidewinder unlocks a notable leap in reliability and scale. They demonstrate clean 5, 10, and 40-fragment assemblies, and benchmark against standard methods where most fail beyond ~5 fragments in comparable conditions. Deep sequencing of a 40-piece construct shows 96.72% valid Sidewinder products, with 100% correct ordering among those reads and 0 misligations across 22,533 observed junctions. The team then stress tests difficult sequence contexts that often break assembly: a GC-rich human APOE coding sequence (up to 95% local GC) assembles with 99.89% Sidewinder products and 100% correct assemblies. A highly repetitive silk segment also assembles successfully, even when multiple fragments share identical toeholds (a condition that would be inherently ambiguous for overlap-driven methods), with residual errors largely attributable to PCR artifacts rather than misassembly.
Finally, they evaluate a pragmatic bioengineering workflow: one-pot parallel construction of multiple distinct genes (dialed out by selective PCR), and high-coverage combinatorial library synthesis. A 10-piece eGFP library with 17 diversified positions (442,368 theoretical variants) achieves 98.88% correct assemblies pre-cloning and a 3WJ misconnection rate of 1 in 960,617, with >91% of theoretical variants observed across sequencing runs.
Overall, Sidewinder broadens the design space – long constructs, extreme sequence regimes, and dense combinatorial diversity become more limited by oligo synthesis quality than by assembly failure, forming the infrastructure that could accelerate synthetic genomics, protein engineering, and AI-driven design-build-test cycles.
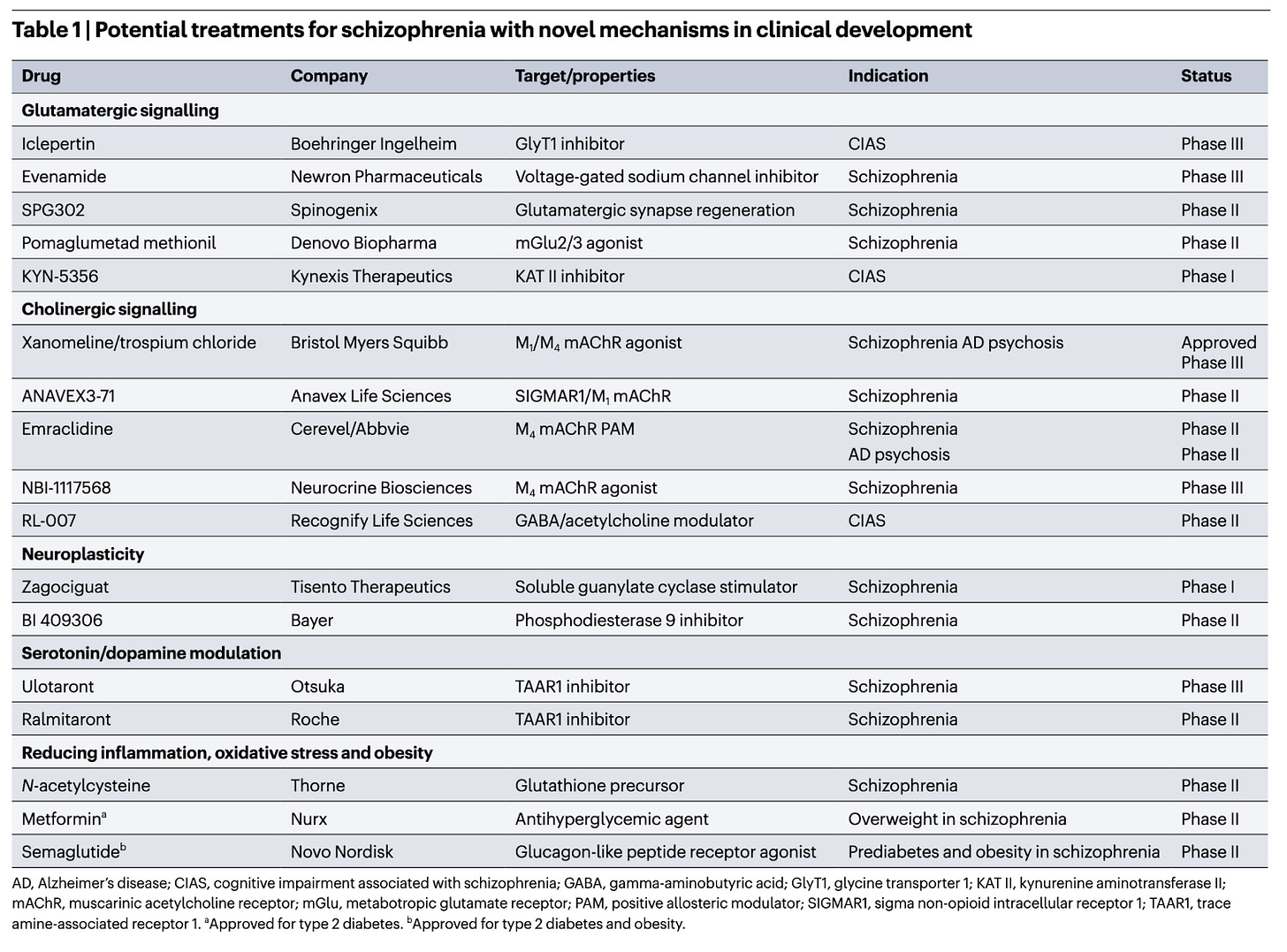
Novel drug treatments for schizophrenia [Coyle et al., Nature Reviews Drug Discovery, January 2026]
Why it matters: Schizophrenia is a complex disorder with many underlying aetiologies; which means a single drug is unlikely to improve all symptoms caused by the disorder. Symptoms are either positive (psychotic, such as hallucinations), negative (anhedonia, social isolation) or cognitive (memory, attention and executive functioning impairment). We need different therapeutic strategies to address the heterogeneity of the disease.
Antipsychotic drugs, most of which act by blocking D2 dopamine receptors, reduce positive symptoms effectively, albeit with a myriad of side effects. They have been ineffective at reducing negative and cognitive symptoms of the disorder.
Breaking a 70 year spell of D2R antagonism as the single mode of treatment, the FDA approved Karuna Therapeutic’s drug in 2024 for the treatment for schizophrenia with no D2R blocking activity, instead using a dual M1/M4 mAChR agonist xanomeline in combination with the peripherally restricted pan-mAChR antagonist trospium chloride.
This review highlights the novel therapeutic strategies in development. Glutamatergic signalling is of special importance, given the substantial representation of glutamatergic risk genes in schizophrenia by targeting NMDARs, KAT II, GlyT1 and mGluR2/3. Acetylcholine receptors (both nAChR and mAChR) have been implicated in the pathophysiology of schizophrenia, and given their reduced expression, their agonism has been suggested as potential therapies. Stimulating neuroplasticity via cGMP, which could improve cognitive symptoms. Zagociguat is currently under development for the treatment of these symptoms via this mechanism. PDE inhibition is an alternative way to increase cGMP. Bayer’s BI409306 Phase II trial didn’t show significant improvement on psychopathology or cognitive improvement.
Notable deals
Andreessen Horowitz and Eli Lilly launch $500M Biotech Ecosystem Venture Fund. In an apparently unprecedented structure, Lilly is providing all the capital while a16z Bio + Health manages the fund. The fund will invest at all stages – from company creation to growth – with a focus on advancing new medicines, enabling novel modality platforms, and scaling emerging health technologies, particularly AI tools. Portfolio companies will also gain access to Lilly’s Catalyze360 program, which provides early-stage biotechs support through Lilly Ventures, Lilly Gateway Labs, and Lilly ExplorR&D. The partnership reflects a broader trend of pharma companies seeking to scout potentially important scientific and technological trends while providing patient capital to enable disruptive companies to realize the full potential of new biological science and AI capabilities.
Isomorphic Labs enters cross-modality, multi-target research collaboration with Johnson & Johnson. The partnership brings together Isomorphic’s AI-first drug design engine, built on AlphaFold 3 and the Nobel Prize-winning protein structure prediction technology, with J&J’s Janssen Biotech unit’s expertise in drug development. Unlike Isomorphic’s earlier deals with Eli Lilly and Novartis (worth nearly $3B combined) which focused primarily on small molecules, the J&J collaboration explicitly extends into large molecule and biologics design, including antibodies, peptides, and molecular glues. Under the terms, Isomorphic will handle in silico predictions and design while J&J conducts experimental assays and assumes responsibility for advancing programs through development. The partnership will target historically difficult-to-drug targets by accessing what the company describes as “planet-scale computing power.” Financial terms and therapeutic categories were not disclosed.
Merge Labs emerges from stealth with a $252M seed round led by OpenAI and Bain Capital to develop non-invasive brain-computer interfaces. The company is pursuing a fundamentally different approach to BCIs than Neuralink’s surgical implants. Merge Labs is developing technologies that connect with neurons using molecules instead of electrodes, transmit and receive information using deep-reaching ultrasound, and avoid implants into brain tissue, aiming to increase neural bandwidth and brain coverage by several orders of magnitude while remaining much less invasive. Valve co-founder Gabe Newell, Bain Capital, Interface Fund, and Fifty Years also participated in the round, which valued the company at $850M. OpenAI has framed BCIs as “an important new frontier” for human-AI interaction and plans to collaborate with Merge Labs on scientific models and bioengineering research.
OpenEvidence closes a $250M Series D led by DST Global and Thrive Capital. The company’s product is an AI-powered medical search engine, built to enable doctors to provide the most informed care based on new publications without needing to spend valuable hours of their day pouring over papers. Unlike other AI models, their multi-AI agentic search engine has only been trained on medical journals and data, improving the reliability and accuracy of the answers it generates. The company itself has grown rapidly, jumping from 3 million monthly consultations a year ago to now 18 million in December 2025. It is currently the most used AI tool by physicians, with 40% using the tool to treat more than 100 million Americans last year alone. Funding from the round will be put primarily towards the necessary compute costs and R&D to advance the product further. This round brings the company’s total raised to nearly $700M and total valuation to $12B.
Think Bioscience raises $55M in an oversubscribed Series A led by Innovation Endeavors, Janus Henderson Investors, and Regeneron Ventures. With their platform, the company seeks to find new druggable locations (e.g. binding pockets) through high-throughput functional surveys. This approach allows them to go after historically undruggable targets with unprecedented success. Think Bioscience’s current lead program targets Noonan syndrome, a genetic condition with a number of detrimental effects including life-threatening cardiac and lymphatic issues, chronic pain, and cognitive impairment. The disease affects approximately one in every 2500 births and has no approved therapies. Other investors in the round were AV8 Ventures, Buff Gold Ventures, CE-Ventures, CU Innovations, MBX Capital, T.A. Springer, and YK Bioventures.
BIOMAKERS closes an $8M round to scale their AI-native platform and expand molecular testing in Latin America. Their platform is an integrated oncology ecosystem which draws on a number of data sources to facilitate both clinical decision making and drug development. Currently operating in 20 countries, the company has amassed a tumor biobank with over 100k samples, high throughput genetic and molecular testing capabilities, and ever-growing datasets of historically underrepresented populations. The round included participation from Endurance28, Labcorp Venture Fund, Latam Impact Fund, Oncology Ventures, Sky High Fund, Sonen Capital, and Zentynel.
In case you missed it
Squidiff: predicting cellular development and responses to perturbations using a diffusion model
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.