BioByte 143: Predicting Structure of Large Protein Complexes, Restoring Naive T-cell Production, Large Scale Reading/Writing of Neuronal Activity, and Valuing Drug Programs with Tripartite Models
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
A Note To Our Readers: This is our last post for 2025 as we will be taking a short break from posting for the holidays. We’ll be back to our regular posting the first full week of January. Thank you from all of us for your support this year and stay tuned for more coming soon, Decoders! Happy holidays! ☃️
What we read
Papers
Scalable prediction of symmetric protein complex structures [Yu et al., bioRxiv, November 2025]
Why it matters: Researchers from the startup EndoFold built Plica-1, a physics-based model that assembles predicted structures of large, symmetric proteins at a fraction of the time previously required. This model unlocks biological inquiry and drug discovery into this class of previously expensive-to-model proteins - including building new drugs that disrupt viral complexes.
AlphaFold has been a breakthrough in protein structure prediction and downstream drug discovery, but it has its constraints. Chief among these is the large memory requirement: the transformers underpinning AlphaFold2/3 require memory that quadratically scales with sequence length (O(L2)), meaning that large proteins and protein complexes do not fit within a single AlphaFold prediction window without significant loss of accuracy or feasibility.
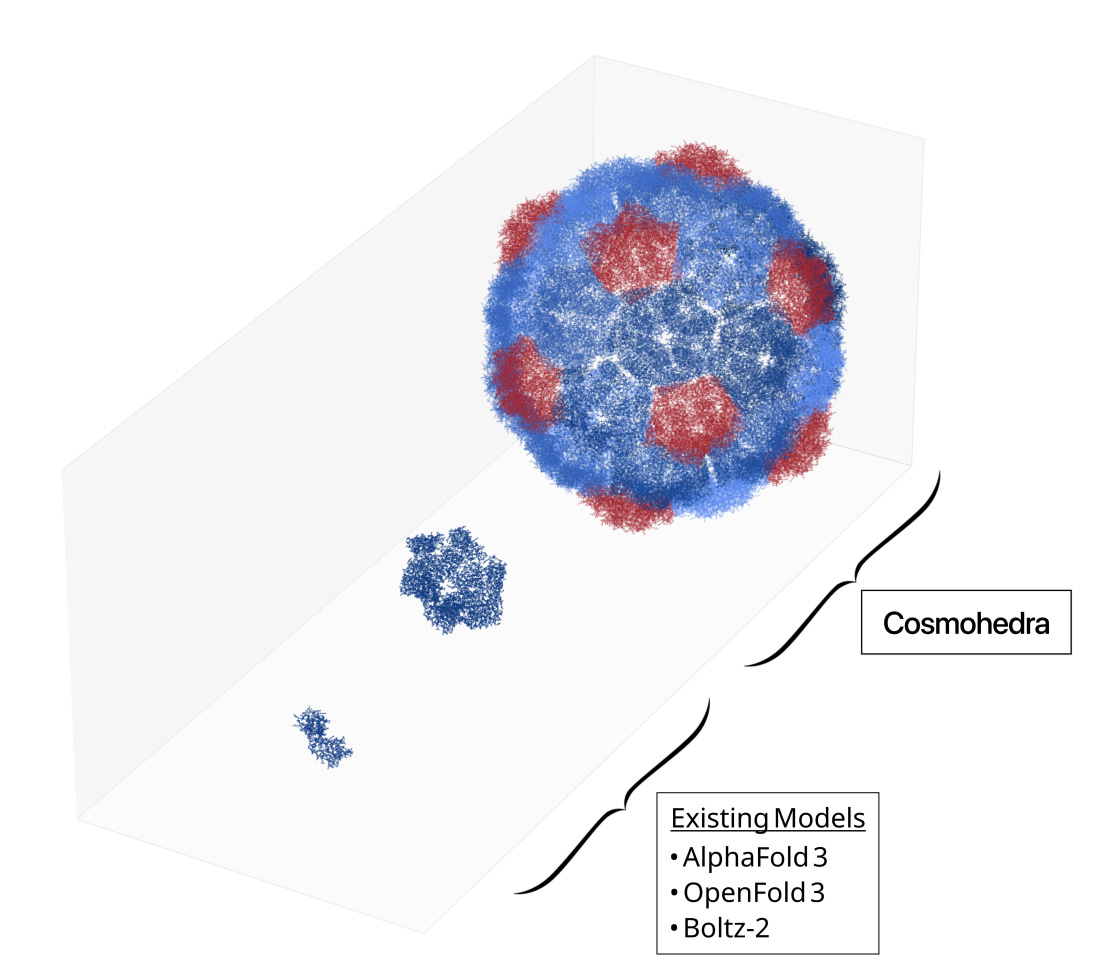
To tackle the problem of structure prediction for large proteins and complexes, the team at EndoFold take advantage of a convenient pattern in biology: protein symmetry. Many large complexes are just symmetric assemblies of smaller monomeric proteins! Take, for example, the Drosophila dArc2 capsid, a retrovirus-like capsid protein co-opted for neuronal cell-to-cell mRNA-based communication. The whole virus-like protein is an assembly of 240 identical Arc2 proteins, each 193 residues large. (Sidenote: Why does nature do this? One explanation is that it is genomically cheaper to encode a single monomer of 579 nucleotides, rather than explicitly encoding a full complex totaling 138,960 nucleotides.)
While AlphaFold can’t model the full complex, it generally can model the monomers of these large, symmetric proteins well! EndoFold builds on this foundation by constructing a physics-based assembly model Plica-1 that uses true and predicted monomer structures and assembles them into symmetric complexes based on their symmetry class. To build the structure of the dArc2 capsid, for example, they assemble the predicted monomer of Arc2 with an icosahedral symmetry. To do this, they built their own geometry-based symmetry library, which enabled a runtime dependence of O(n log n), which is a huge upgrade over O(n2) for large proteins. Previous methods, such as docking ones like EvoDOCK, often require tens of GPU hours to predict complexes on the order of a few hundred residues. By their estimates, Plica-1 to their estimate is 103-105x faster - predicting large complexes in tens of seconds.
Given relatively accurate monomeric structures, Plica-1 assembles proteins with resolution within range of the experimental measures themselves. The experimental methodologies to develop structures for large complexes, such as cryoEM and X-ray crystallography, aren’t perfect - they have noise within the structures on the order of 1-5Å. On a set of 36 symmetric complexes predicted from sequence, Plica-1 achieves RMSDs within that range of 1-5Å. This means that it’s actually difficult to tell whether the Plica-1 structures or the original cryoEM structures are more accurate to the true underlying protein structure!
One acknowledged limitation of Plica-1 is that it inherits whatever biases and errors exist in the monomeric structures it assembles. If a monomer is misfolded or lacks the correct interface geometry, no amount of downstream assembly can fully recover the true complex. But this dependency also points to a broader opportunity. By turning high-quality monomer predictions into near-experimental complex structures at scale, Plica-1 effectively acts as a synthetic data generator for large protein assemblies. Rather than competing with end-to-end structure predictors, it complements them by enabling rapid exploration of complex architectures, guiding experimental design, and potentially seeding the next generation of complex-aware foundation models. EndoFold is already testing this paradigm in a campaign to inhibit Bufavirus 1 capsid formation, using a predicted macrocyclic binder designed against a single capsid monomer. More broadly, the speed and accuracy of Plica-1 open the door to systematic study and therapeutic targeting of large, symmetric protein complexes that were previously out of reach.
Transient hepatic reconstitution of trophic factors enhances aged immunity [Friedrich et al., Nature, December 2025]
Why it matters: Friedrich et al. detail a novel approach using mRNA-LNPs to reprogram liver cells to compensate for age-related decline in T cell generation capabilities in the thymus. This treatment restores naive T cell production and dendritic cell function, significantly boosting vaccine responses and antitumor immunity. This method offers a safe, reliable avenue for preserving immune resilience in elderly mammal populations.
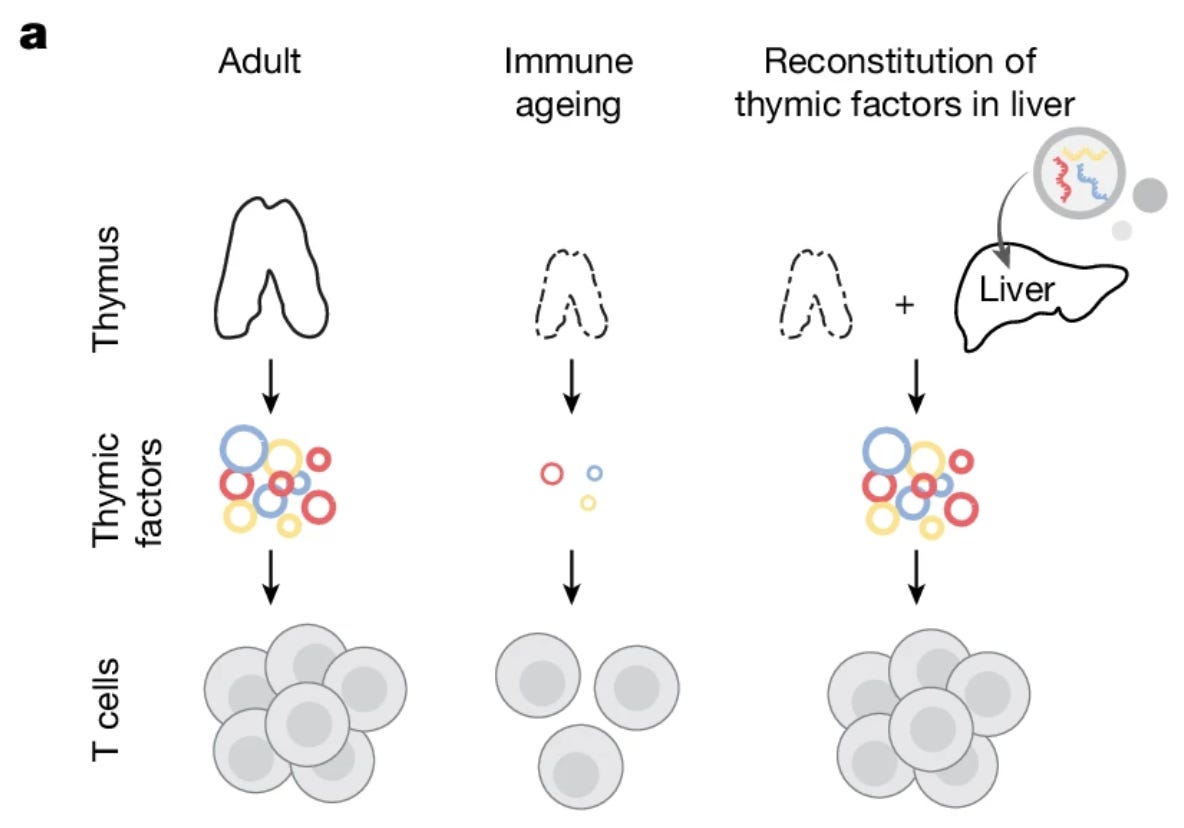
The thymus is responsible for producing T cells that play a central role in the overall immune system’s function; as humans and most other mammals age, the thymus shrinks (medically referred to as involution), causing decreased naive T cell output, less TCR repertoire diversity, and generally weakened primary responses. While conventional methods to combat the age-related decline have focused on the administration of hormone, cytosines, and small molecules, such approaches suffer from limited effect size, toxic side effects, and generally suboptimal clinical feasibility. In this work, the authors describe an alternative approach centered around “reconstituting thymus-derived factors in the liver.”
The authors began with the goal of identifying potential T cell pathways for targeted restoration of immune function. They began by probing trophic (nutritional) signalling mechanisms in mice of various ages using single-cell RNA sequencing and TCR-seq along with spatially resolved information. The resultant datasets, spanning mice at birth up to 90 weeks of age, revealed shifted T cell states, such as the loss of stem-like populations, expanded memory states, and exhaustion-like phenotypes. Comparing the multi-omic mouse data, the team identified three crucial signaling pathways that showed consistent decline with age - Notch signaling (DLL1) known to be crucial for the initial commitment of stem cells into the T cell, FLT3 ligand (FLT3L) which is important for hematopoietic stem cell expansion and immune cell development, and finally, Interleukin-7 (IL-7) which is essential for T cell maturation and survival. These three targets, collectively referred to by the team as DFI, formed the basis for a potential approach to recover superior immune signalling in older mice. Notably, the authors chose DLL1 over other trophic factors like DLL4 for better safety characteristics since the latter can restrict lymphoid differentiation due to its role in angiogenesis and vasculature remodeling. Rather than targeting the signaling pathways in shrinking thymuses, the authors chose to “[reconstitute] the identified signalling pathways ectopically in the liver,” due to the greater ease of priming and maintaining adaptive immunity and the enduring protein-synthesis capabilities across a mouse’s lifespan. The team chose to develop LNP packaged mRNAs encoding the three proteins rather than use recombinant proteins due to the minimal immune side effects and optimizations in mRNA stability and delivery. The LNP cocktail primarily targeted the liver, where all three DFI mRNAs were confirmed to show robust translation in hepatocytes. Before moving on to more focused experiments, the authors assessed the safety of the DFI approach, confirming that all three factors remained bioavailable in the liver and there were no aberrant effects in the way of weight gain, decreased liver function, or significant inflammation.
The researchers then evaluated the therapeutic potential of the DFI reconstitution approach through three experiments: effects on immune ageing phenotypes, vaccine response, and antitumor immunity. First, they observed that DFI reverses the characteristic “memory bias” of aging by restoring naive T cell counts and de novo thymopoiesis through the expansion of common lymphoid progenitors. This rejuvenation extended to the broader immune environment, where the therapy expanded cross-presenting cDC1 dendritic cells and reduced pro-inflammatory, age-associated B cells in the spleen. In vaccination trials, DFI pre-treatment doubled the frequency of antigen-specific CD8+ T cells, effectively restoring the immune responsiveness of aged mice to levels seen in young adults. When applied to aggressive melanoma models, the therapy worked synergistically with immune checkpoint blockade mechanisms to drive robust T cell infiltration and complete tumor rejection in 40% of cases. Importantly, the liver-based mRNA delivery ensured that these trophic signals were produced transiently, avoiding the systemic toxicity and inflammation often associated with recombinant protein therapies. In summary, the work presented in the paper demonstrated the potential of mRNA-based therapeutic platforms to create safe avenues for the restoration of biological function and potentially aid broader longevity related research.
Large-scale cellular-resolution read/write of activity enables discovery of cell types defined by complex circuit properties [Drinnenberg et al., bioRxiv, October 2025]
Why it matters: All-optical neuroscience has matured from single-cell demonstration to reliable control of dozens of cells. However, key questions about how distributed neural ensembles shape downstream dynamics requires simultaneous, cellular-resolution control and dense population readout across millimeter-scale tissue. To bridge this gap, the authors offer two advancements: (a) efficient, soma-targeted molecular tools (a new soma-enriched excitatory opsin line based on ChRmine together with soma-localized GCaMP variants) and (b) a hybrid ‘follower-focused’ experimental design, where opsin expression is restricted to a targeted subpopulation while a dense, pan-neuronal calcium indicator (ribo-GCaMP8s) labels the broader circuit. With this approach, the authors obtain an order of magnitude larger scale in optical read/write capabilities: reliable stimulation of ~1,000 targeted neurons while reading out activity from ~10,000 neurons across a 1x1 mm field – with tight spatial precision, low per-cell power, minimal cross stimulation (unintended opsin activation by the imaging laser), and long-term stability. This leap turns all-optical experiments from constrained, small-N manipulations into systematic, causal perturbation studies that can reveal circuit motifs only visible at scale.
In experiments targeting the visual cortex of mice, the team stimulated ~50 excitatory neurons (‘ensembles’) – with 10-12 ensembles per mouse – while monitoring the surrounding network. Their dense, downstream readout revealed a reproducible subpopulation of non-targeted neurons – GER (general ensemble response) cells – that were reliably activated by many, distinct stimulation ensembles (recruited by ≥4 different, non-overlapping ensembles far more often than chance). Modeling and experiments show GER cells are predominantly somatostatin-expressing (SST) interneurons in deeper cortex strata (Layers 2/3), forming a previously uncharacterized subtype. These cells have broad stimulus responsiveness, high pairwise synchrony, and integrate over long spatial ranges; their recruitment is inconsistent with naive spatial random-connectivity models. Practically, that means a previously hidden, inhibitory subnetwork acting as a broad feedback mechanism – one that can monitor and normalize distributed excitatory activity across hundreds of microns – a circuit role that could only be exposed by the new scale of perturbation and readout enabled by this effort. Ultimately, this paper establishes a robust methodology for not only interrogating causal links between neural ensembles and behavior, but for discovering and characterizing cell types defined by their functional roles beyond molecular markers.
Tripartite models for estimating the value of drug candidates and decision tools [Mellnik & Scannel, arXiv, March 2025]
Why it matters: Valuing drug development programmes is an ongoing challenge in the pharmaceutical industry. The commonly used risk-adjusted net present value (rNPV) approach has significant limitations that can lead to suboptimal investment decisions. Mellnik & Scannel introduce the “tripartite model” as a solution to these limitations; by linking the financial value of the programmes with the quality of the ‘decision-making tools’ used in development.
Where do classical rNPV valuation approaches fail?
rNPVs are point estimates, which fail to capture the range of potential outcomes. Therefore it doesn’t accurately convey the risk associated with a drug development programme. Monte Carlo simulations can be helpful to estimate risk, but it is challenging to accurately estimate probabilities of rare events (such as developing a blockbuster drug).
They are limited by insensitivity to decision quality at each step in the R&D process (that is to the predictive validity of the decision tools used to decide which candidates should progress to the next stage and which ones should be terminated early). The ‘probability of success’ used in rNPV calculations is actually the stepwise progression probability (Pn) which is independent from future progression probabilities (Pn +1), and therefore Pn+1 doesn’t capture upstream decisions.
The tripartite model proposed by the authors can value the candidates, decision tools used, and the interactions between the two. The model contains three levels of information: the observed result of a decision (δ̂; assay readouts, biomarker readings, clinical endpoints), the unobserved ground truth of a decision tool (δ; true value of decision tool without measurement errors), and the ground truth capacity of the drug candidate to achieve commercial and technical success (G; treats commercial success as a continuum rather than binary outcome). The relationship between δ and G is modeled using a bivariate normal distribution, where the correlation coefficient ρ represents the “predictive validity” of the decision tool.
Multiple tripartite models can be placed in series to describe a multi-stage drug development process. Each stage takes into account changes in decision tools between stages. By using these models in series, changes in the predictive validity of decision tools used in early stages allow the modeller to see how it affects the value of the candidate in later stages.
Notable deals
Sanofi has agreed to license ADEL’s Alzheimer’s drug candidate for $80M upfront, with the potential for up to $1.04B contingent on developmental and commercial milestones. ADEL’s drug, ADEL-Y01, is a humanized monoclonal antibody designed to inhibit the aggregation and propagation of toxic tau protein. This approach stands in contrast to other drugs which target total tau, as it utilizes the acylation of lysine-280 to distinguish between the toxic and healthy forms of the protein. Since 2020, the South Korean biotech has developed the asset in partnership with Oscotec, another South Korea-based pharma, who is eligible for 47% of the proceeds. ADEL-Y01 is currently in Phase I clinical trials, of which Phase Ia results were recently shared earlier in December.
Chai Discovery has raised $130M for their Series B, more than doubling their previous valuation of $550M to $1.3B. This round—led by General Catalyst and Oak HC/FT—follows their $70M Series A in August. It also comes shortly after the release of their recent preprint on bioRxiv, which demonstrated the ability of Chai-2 to design drug-like, monoclonal antibodies with strong developability profiles. Other investors in the round included Dimension, Emerson Collective, Glade Brook, Lachy Groom, Menlo Ventures, Neo, OpenAI, SV Angel, Thrive Capital, and Yosemite.
Ambros Therapeutics announced their launch this week with a $125M Series A. Funding will be used to develop neridronate—a non-opioid pain medication—through Phase III trials for Complex Regional Pain Syndrome Type 1 (CRPS-1) with Breakthrough Therapy, Fast Track, and Orphan Drug designations. The candidate itself was licensed from the original developer Abiogen Pharma, and has been used to treat 600k patients to date since its approval in Italy in 2014. As per their agreement, Ambros is granted exclusive licensing rights to the drug in North America, with the potential for expansion into other markets. The round was led by Enavate Sciences and RA Capital Management, with additional support from Abiogen Pharma, Adage Capital Partners LP, Arkin Bio, Balyasny Asset Management, Janus Henderson Investors, and Transhuman Capital.
The Stanford spinout Link Cell Therapies has launched with a $60M Series A, seeking to specifically target cancerous tumors with CAR-T therapies. Link has built its platform on logic-gated CAR-T cell control, a technology developed at Stanford which facilitates specificity by only activating and killing cells when specific antigen combinations are in close proximity. The antigen combinations are selected for their uniqueness to the cancerous cells, avoiding potential destruction of healthy cells. Link’s lead program, LNK001, is on track for IND approval and the start of Phase I for renal cell carcinoma sometime in 2026, with a second program for colorectal cancer planned for Phase I in 2027. The round was led by Johnson & Johnson Innovation with participation from Samsara BioCapital, Sheatree Capital, and Wing Venture Capital, among others.
Aether Biomachines has raised $15M, led by Tribe Capital. The funds will be used to improve their capabilities in designing and optimizing protein nanofactories capable of synthesizing next-gen materials with atomic precision, while also allowing the company to scale existing products and pursue further applications across manufacturing, national security, and sustainability. Aether has already succeeded in designing additives for 3D printing, affording 10x faster print times and 2x strength, as well as new classes of proteins capable of extracting rare earth metals, capturing carbon, or breaking down forever chemicals. Other investors in the round included 4DX Ventures, Henkel Corporation, Natural Capital, Radicle Impact, Resilience Reserve, Shrug Capital, and Unless Partners.
In case you missed it
Neuroscientist Doris Tsao joins Astera to lead its new neuroscience program
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.









Hi Mikaela ...the liver reprogramming hack you mention here... the Friedrich et al. paper... it is visceral. It is not "healing" in the poetic sense. It is routing around a failed node. The thymus bottlenecked. So they forced the liver to pick up the slack. That is how you fix a system that is too old to fix itself. You do not ask the broken part to work harder. You build a bypass.
And this Tripartite Model for valuing drug programs. Linking value to the "predictive validity" of the decision tool. That hits hard. We value the output (the drug. the treaty). We ignore the tool that got us there. If the tool has a 30-year latency... the value of the output is effectively zero.
So here is the question. We are building O(n log n) speed for proteins. We have optical read/write for brains. But we are still using low-fidelity, high-latency tools to decide which global problems to solve. Do we need a Tripartite Model for planetary survival... one that penalizes the "decision tools" (like bureaucracy) that are too slow to matter?
The first episode is live here... https://silentwitnessin.substack.com/p/file-001-the-thermodynamic-bluff?r=6r3orq
(My new serialized novel, The Man Who Blocked The Sun, follows a diabetic engineer who hacks the climate because the treaties are too slow. It'd mean the world to me if you'd join to follow the work.)
Appreciate the breakdown on tripartite models here. The idea of explicitly tying decision tool quality to candidate value thru the correlation coefficient ρ makes alot of sense, especially when you consider how many pharma decisions fail not becuase of the candidate but the predictive validity of assays at each stage. I saw something similar play out at a biotech I was advising where early stage assays had good throughput but weak correlation with Phase II endpoints, and that mismatch basically created false confidence in the pipeline. Curious if there are practical limits to estimating ρ accurately in real scenarios, since rare event modeling is already dicey and layering in tool validity might amplify noise.