BioByte 137: C2S-Scale Parses Single-Cell Data into Sentences, Stabilizing TRegs for Personalized Cell Therapies, Quantum-augmented NMR, and How the Powerhouse of the Cell Also Dictates Sleep
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Papers
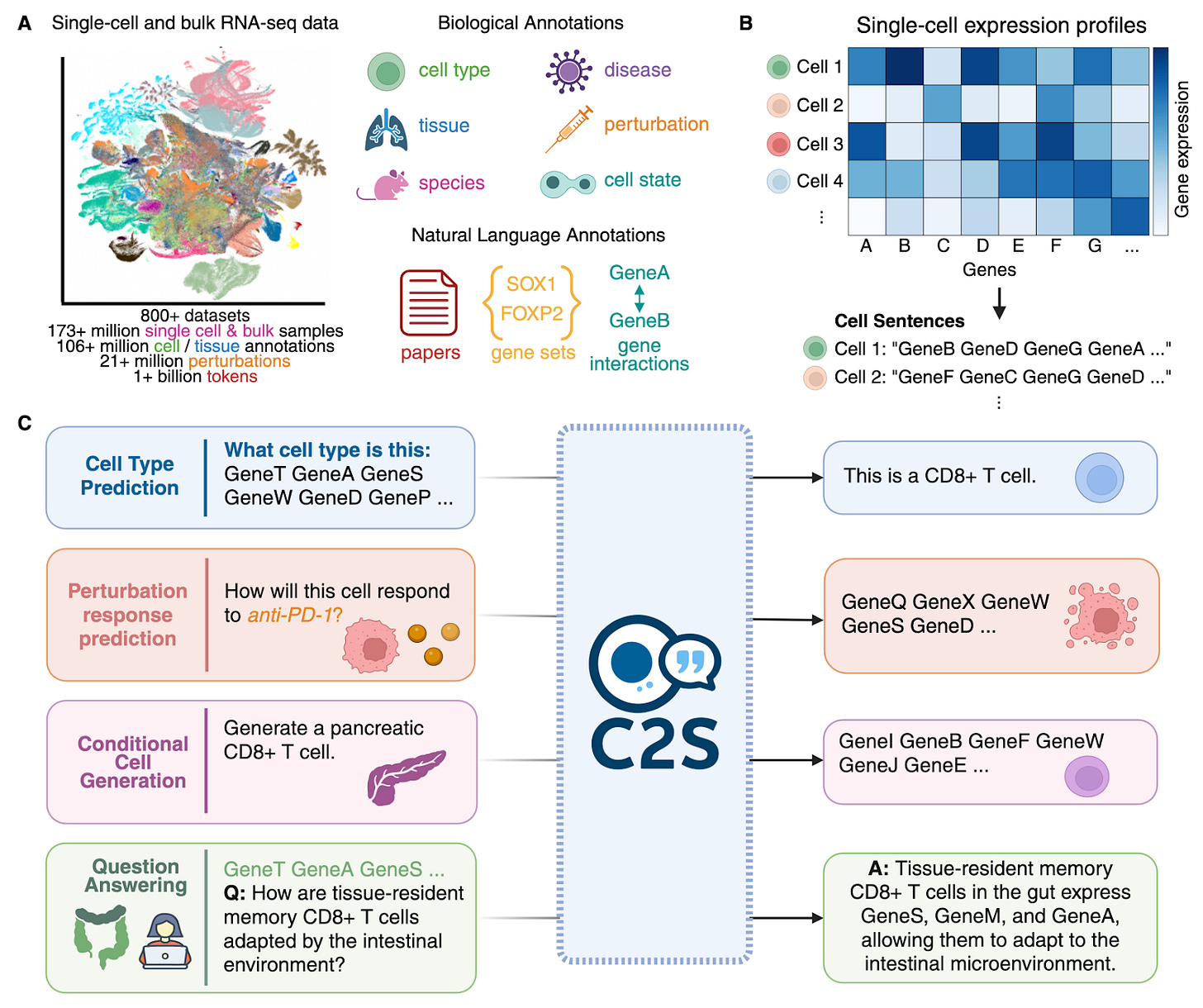
Scaling Large Language Models for Next-Generation Single-Cell Analysis [Rizvi et al., bioRxiv, October 2025]
Why it matters: In this paper, a joint team of researchers across Yale, Brown, USC, and Google present the Cell2Sentence-Scale (C2S-Scale) model family. Building on their previous work of Cell2Sentence, C2S-Scale uses large language model frameworks to represent single-cell RNA sequencing as “sentences” of gene expression data along with additional modalities like biological text and metadata. After fine-tuning with reinforcement learning techniques, C2S-Scale demonstrates promising performance across a variety of tasks including perturbation effect prediction and more general biological reasoning tasks.
Most presently available machine learning models for handling scRNA-seq data rely on bespoke architectures that are unable to utilize additional natural language data modalities for predictive and generative tasks. Cell2Sentence (C2S), the precursor to C2S-Scale released in 2023, showed that LLM architectures could be used to reason over single-cell data converted into a sentence-like structure. Specifically, C2S represented genes and their expression in decreasing order like a “textual sentence”, with the authors finding that such a representation seemed to be “reversible with minimal information loss due to the strong relationship between relative position and original gene expression.” The C2S-Scale family of models builds on top of the C2S design with separate pretraining and finetuning across a range of tasks like single-cell and multi-cell conditional generation and annotation. Specifically, the models were pretrained on over 50 million human and mouse transcriptomes, with additional natural language data in the form of annotations and papers, allowing them to capture relationships between genes and relative expression as well as respond to prompts about the data.
C2S-Scale was evaluated against both standard scRNA-seq tools as well as conventional natural language models to test associated reasoning capabilities. The models achieved comparative results with expression-only foundation models, with the authors noting that performance on matching single-cell data profiles with bulk data indicated “that C2S captures a more biologically meaningful representation of cellular states through cell sentences.” C2S-Scale was also compared against models like Llama, Gemini, and GPT-4o, demonstrating superior performance in description and summarization tasks at both the individual cell level, cluster level, and whole dataset level. Interestingly, when training various sizes of C2S-Scale, the authors observed scaling law behavior, with larger models showing consistently better performance across cell-type annotation, tissue inference, and conditional generation tasks, even for parameter efficient fine-tuned versions. Furthermore, while the cell sentence input representation did not inherently contain spatial information, C2S-Scale was able to recapitulate such relationships through cell neighborhood prediction and generation tasks.
Finally, C2S-Scale was also put through a perturbation response prediction test, where the models were tasked with predicting gene expression patterns under certain perturbation conditions. This scheme was paired with Group Relative Policy Optimization (GRPO) reinforcement learning to optimize specified “key gene programs of interest” like apoptosis and interferon response mechanisms based on the perturbation dataset. The authors also introduced an additional metric that they called the single cell Frechet Inception Distance or (scFID) which measures higher order variation across different cell-state embeddings. On both scFID and other conventional metrics, C2S demonstrated competitive performance on perturbation response, especially on fully unseen combinatorial perturbations. To demonstrate C2S’s real-world utility, the authors configured the model to run a “dual-context” virtual screen to identify drugs that could increase immune visibility by boosting the MHC-I antigen-presentation program. Specifically, C2S was presented with one setting where cells had low (but still significant) levels of interferon signaling and another with no activity. After predicting the effects of various drugs, the model identified silmitasertib as a potential candidate that could significantly increase interferon signaling without affecting the neutral state cells. Despite there being no such record of this activity pattern in existing literature, wet-lab validation of the drug in neuroendocrine cells confirmed the in silico hypothesized pattern of activity.
C2S’s ability to integrate natural language techniques into single-cell modeling paradigms is interesting for multiple reasons. Apart from potentially learning more expressive cell-state embeddings and providing a promptable interface to reason over literature and data, the emergence of scaling laws lead to questions of how bigger models may potentially lead to additional hypotheses that can be validated experimentally. C2S-type frameworks might also be interesting to pair with additional data like proteomics and clinical records, potentially opening the door for more patient-specific conclusions.
Mitochondrial origins of the pressure to sleep [Sarnataro et al., Nature, July 2025]
“Sleep, like aging, may be an inescapable consequence of aerobic metabolism.”
To understand the molecular changes that underpin the need for sleep, this team compared the transcriptomes of single neurones of rested and sleep-deprived flies. They found that transcripts upregulated after sleep deprivation, in sleep-regulating dorsal fan-shaped body neurons (dFBNs) but not in the rest of the brain, encode for proteins with roles in mitochondrial respiration and ATP production.
The expression of these genes accompany multiple morphological changes: “mitochondrial fragmentation, enhanced mitophagy and an increase in the number of contacts between mitochondria and the endoplasmic reticulum, creating conduits for the replenishment of peroxidized lipids.” These changes revert after sleep. The results show that the induction or prevention of mitochondrial fusion or fission in dFBNs alters sleep and the electrical properties of sleep control cells: fused mitochondria increase and fragmented mitochondria decrease neuronal excitability and sleep.
During waking, arousal inhibits dFBNs, its ATP consumption decreases and therefore ATP accumulates. This increases electron leakage to O2, leading to reactive oxygen species (ROS) risk. During sleep, dFBNs are active and thus consume ATP, and the proton-motive force is spent due to the CoQ pool being oxidized. Through mitochondrial interventions in dFBNs, the authors showed that this redox mismatch is causal for sleep pressure. By providing an alternative path for excess electrons, sleep need is reduced. Conversely, using a light-driven proton pump raises ATP without reducing NADH and precipitates sleep. This shows that the circuit for sleep homeostasis in flies emerges from a metabolic imbalance in a specific, defined sleep circuit.
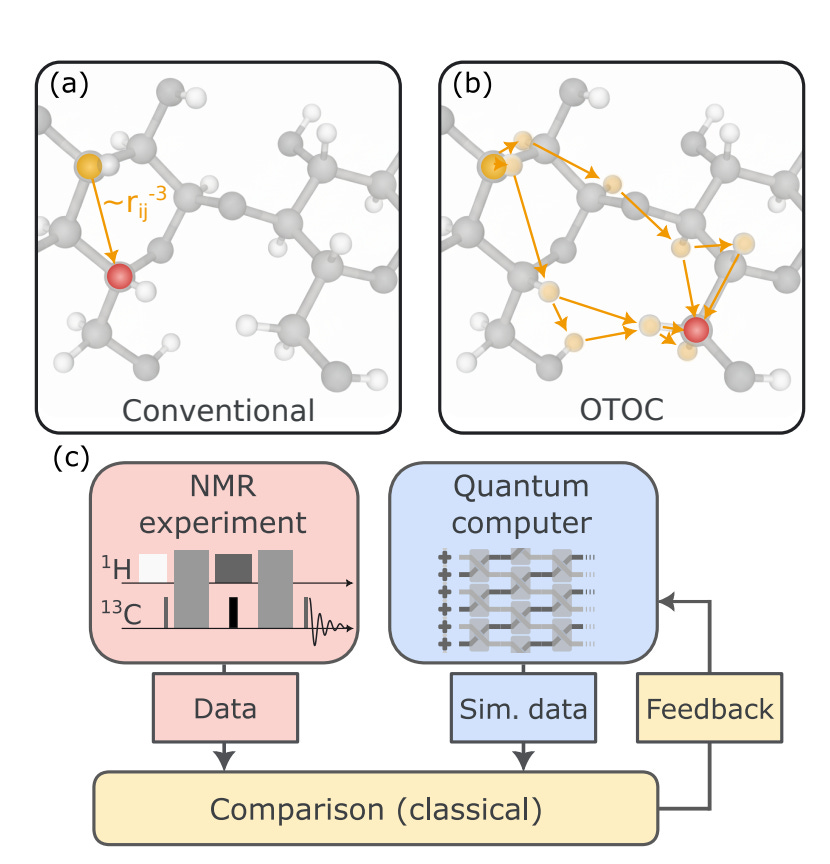
Quantum computation of molecular geometry via many-body nuclear spin echoes [Zhang et al., Google Quantum AI, October 2025]
Why it matters: Nuclear magnetic spectroscopy (NMR) is one of the few experimental windows into molecular structure and behavior in solution, but conventional NMR observables – including chemical shifts (measuring local electronic environments), scalar J couplings (reflecting bond connectivities), and NOEs (identifying molecular contacts spatially) – provide short-range, pairwise geometry. Thus, ambiguous and flexible conformations remain notoriously difficult to analyze. Zhang et al. address this by measuring many-body nuclear spin echoes with out-of-time-ordered correlators (OTOCs) – a correlation function for how a local perturbation ‘scrambles’ and spreads through a network of interacting spins – and interpreting them with compact quantum simulations. By doing so, they extract long-range, high-order correlation information – revealing how multiple parts of a molecule collectively interact. In other words, OTOCs are a quantum analog of the butterfly effect, and can only be feasibly assessed with quantum circuits. In the near term, that means tighter, less ambiguous conformational ensembles for small-to-medium molecules: better stereochemistry assignment, more reliable conformer populations for medicinal chemistry, and fewer failed structural hypotheses in lead generation. Long-term, if the approach scales, it promises a qualitatively new spectroscopy. A quantum-augmented NMR could supply rich, many-body priors to ML or molecular dynamics (MD) pipelines, enabling structural determination of flexible motifs and complex assemblies that are inaccessible or require massive orthogonal datasets today.
The paper stitches three elements into a working loop. First, they record OTOC time traces in molecular samples (small organics in liquid crystal environments) – an observable measuring multi-spin scrambling rather than only pairwise couplings. Second, because classical simulation of those many-body correlators is expensive, the team compiled shallow, hardware-efficient circuits and ran them on a near-term quantum processor. They carefully organized the operations to minimize error accumulation and execution time (circuit scheduling), and applied an error-mitigation protocol (Pauli-pathing zero noise extrapolation) to estimate the quantum measurement as if there was no hardware noise, recovering credible time traces of many-body spin correlations. This can be framed as a verifiable quantum-assisted measurement pattern: select an experimentally accessible but classically infeasible observable, simulate it on a quantum device with provably tractable circuits, and reduce noise so that the quantum output becomes a trustworthy surrogate for the true many-body signal. Finally, they folded the measured and simulated traces into molecular modeling – constraining MD/force-field ensembles – and validated reconstructed distances and dihedrals against independent spectroscopic references with minimal errors in their test cases.
The advance is pragmatic, testable, and promising given it does not yet require fault-tolerant quantum computers. However, the currently demonstrated molecules are small (systems of tens of nuclear spins) and measured in controlled liquid-crystal-like environments. Key priorities to realize the promise of the approach include higher-fidelity quantum processors, improved OTOC signal-to-noise ratio in realistic biological samples, and standardized pipelines that translate OTOC constraints into ML/MD priors. If the engineering milestones are met, the payoff is concrete: a new class of spectroscopic priors that tames critical structural problems (hard-to-crystallize motifs, flexible conformational ensembles, ambiguous stereochemistry), materially reducing the experimental burden in drug discovery and structural biology.
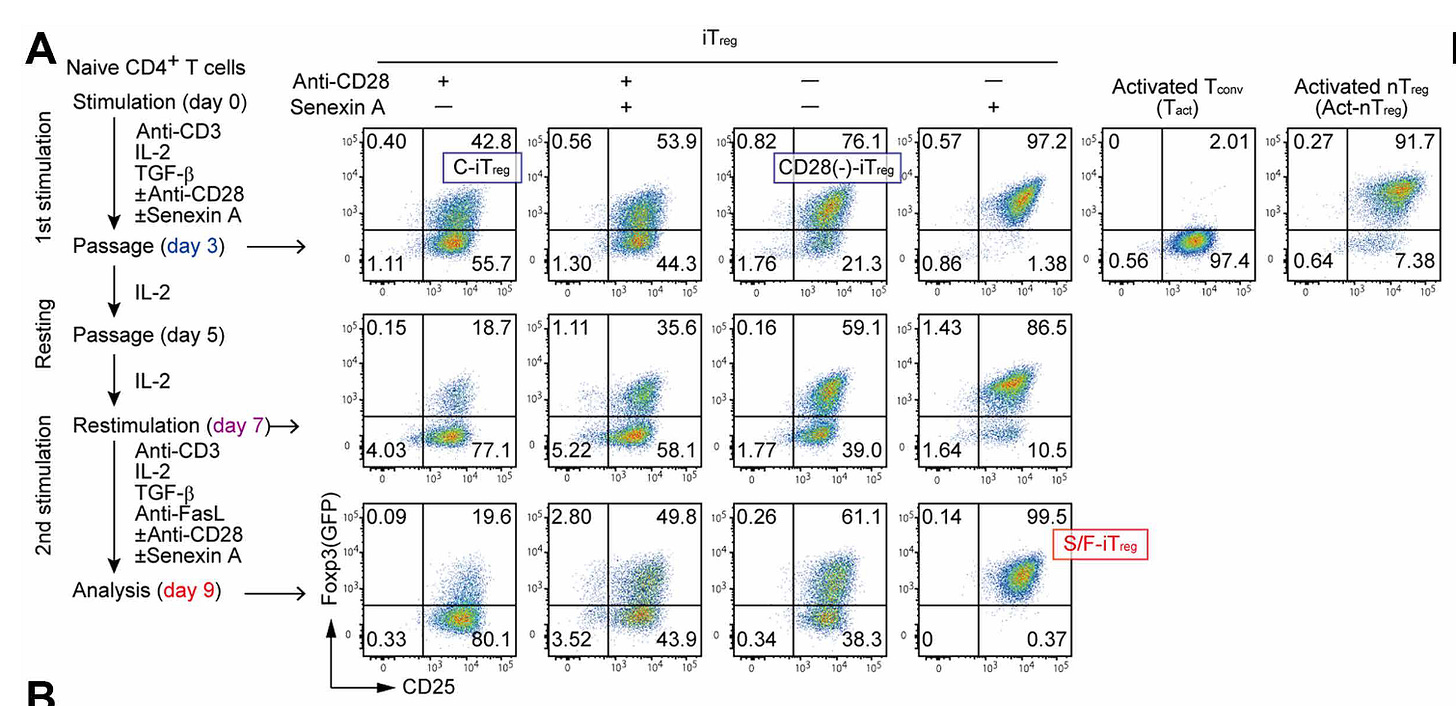
Generating functionally stable and antigen-specific Treg cells from effector T cells for cell therapy of inflammatory diseases [Mikami et al., Science Translational Medicine, October 2025]
Why it matters: Regulatory T-cells (Tregs) are key components in maintaining immune tolerance through the dampening of inflammation, and there’s increasing promise in using them as personalized cell therapies for autoimmune and inflammatory disorders. However, natural Tregs (nTregs) extracted from patients’ blood are difficult to expand and maintain in vitro. On the other hand, efforts to derive Tregs in vitro from patient conventional T-cells (Tconv) have resulted in Tregs without long-term, stable immune suppression. Mikami et al. developed a strategy to stably convert Tconv into Tregs with long-lasting suppressive activity, opening up a path toward manufacturing patient-specific Tregs for cell therapy.
Treg cells expressing the transcription factor Foxp3 and surface markers CD25 and CTLA-4 are central to controlling immune overactivation. They suppress inflammatory T cells and help maintain tolerance to self-antigens. Traditionally, methods to induce Treg cells have involved TGF-β and IL-2 to express Foxp3, but time has held that the TGF-β - mediated induction of Foxp3 fails to drive long-term expression, due to lack of a stable epigenetic signature found in natural Tregs. In high inflammatory contexts, these induced cells lose Foxp3 expression and revert back to inflammatory T-cell phenotypes. Small-molecule based methods of Treg induction also lead to the same problem, through the same channel - activating Tregs through TGF-β - mediated Foxp3 upregulation. As an analogy, the Foxp3 switch was flipped on, but without an epigenetic “lock”, strong immune contexts flipped the switch back off.
To solve this, the authors combined two mechanistically distinct interventions that target these two layers of Treg identity. First, to induce Foxp3 expression without using TGF-β, they used a CDK8/19 inhibitor (senexin A) to upregulate Foxp3 through enhanced STAT5 signaling (switch is on). Second, they removed CD28 co-stimulation during T-cell activation - a condition previously shown to trigger DNA hypomethylation at Treg-specific enhance regions such as Foxp3 CNS2, Ctla4, and Il2ra (lock is on). Together, these steps enabled both Foxp3 induction and installation of Treg-type epigenome, enabling these induced Tregs to stay Tregs. The success of this approach underscores a broader principle: durable cell therapies require attention to both transcriptional programming and epigenetic remodeling, not just transient gene expression.
To demonstrate that this works, Mikami et al. generated what they call stable/functional iTregs (S/F-iTregs) from both naïve and effector/memory T cells in mice, and even from human peripheral T cells. These S/F-iTregs nearly completely converted to a Foxp3+ state, possessed nTreg-like chromatin accessibility and hypomethylation, and maintained its suppressive function in inflammatory environments. In mouse models, they prevented inflammatory bowel disease (IBD) and mitigated graft-versus-host disease comparably to natural Tregs. Crucially, the approach also worked on human cells from healthy donors and lupus patients, highlighting its clinical potential towards manufacturability of stable, personalized immunosuppressive cell therapies.
Notable deals
Adaptyx emerges from stealth with $14M seed led by Interlagos. Originally spun out of Stanford and the Chan Zuckerberg Biohub, the company develops a myriad of biowearables powered by programmable molecular switches which consist of highly sensitive lab-made DNA bioreceptors which bind to target molecules. Adaptyx seeks to address the data deficit in healthcare, ushering in an age of continuous monitoring rather than the moment-in-time snapshots of health yielded by current lab tests. Applications are wide for this technology with those currently in development encompassing heart failure management, hormone optimization, care delivery augmentation via real-time monitoring of drug levels and critical care biomarkers, and personalized wellness which can give insights into stress, metabolism and aging allowing for proactive and preventative health choices. The seed funding raised will be used to accelerate R&D and product development, expand the pipeline, and support new hiring as the company scales. Other investors in the round include: Overwater Ventures, Starbloom Capital, Stanford University, the Chan Zuckerberg Biohub, Hyperlink Ventures, Cantos Ventures, Humba Ventures, and Seaside Ventures.
Electra Therapeutics raised a $183M Series C round led by Nextech and EQT Life Sciences. The funding will go towards the global Phase 2 and 3 of their lead candidate ELA026, exploration of ELA026 in other hematologic cancers, and the development of ELA822—their second pipeline program—into the clinic. Electra seeks to address secondary hemophagocytic lymphohistiocytosis (sHLH) with ELA026, a disease which has a 50% survival rate after two months with current treatments. The drug has demonstrated to be promising, as in their Phase 1b study, ELA026 increased the two month survival rate to 100%. Their other program, ELA822, seeks to deplete activated T lymphocytes, with applications in diseases across immunology and inflammation. This round is a significant increase from their $84M raise in 2022.
Takeda partners with Innovent Biologics in a deal worth up to $11B to strengthen their oncology pipeline. The deal nets Takeda rights to the development, manufacturing, and commercialization of three oncology drugs outside of Greater China: two late-stage (IBI363 and IBI343) and one early-stage (IBI3001). IBI363, a PD-1/IL-2α-bias bispecific antibody fusion protein, is currently being evaluated for non-small cell lung and colorectal cancer, while IBI343—an antibody-drug conjugate (ADC) targeting Claudin 18.2—is being applied towards gastric and pancreatic cancers. IBI3001 is a bispecific ADC that targets EGFR and B7H3 to treat advanced or metastatic solid tumors. With the deal, Innovent receives $1.2B upfront, with the remainder falling into milestones and royalty payments. This deal highlights the continuing trend of global pharma companies shopping for new therapeutics out of China’s rapidly advancing position as a biotech stronghold.
Ampa raised $8.5M in pre-Series A funding led by Nexus NeuroTech Ventures. Emerging from stealth in June 2025, Ampa is seeking to implement transcranial magnetic stimulation (TMS) for treatment of major depressive disorder (MDD). With depression rates skyrocketing in the last several decades, CEO Don Vaughn professes the need for greater treatment optionality besides medication, which often come with a long list of significant side effects. TMS is by no means a new treatment and has been established as a highly effective therapeutic modality for MDD, as well as a host of other neurological and neuropsychological disorders. The technology, however, has long been limited by difficulty in accessibility due to expensive and cumbersome equipment, complexity of administration, and logistics and timing constraints surrounding intensive treatment regimens. Ampa has developed a portable, camera-guided version in their Ampa One product which achieved FDA clearance in February of this year and has initiated rollout in the US. The platform is subscription-based and integrates seamlessly with telemedicine, allowing patients to complete therapy in the comfort of their own homes. Other participating investors include: Satori Capital, Morningside Ventures, Continuum Health Ventures, and the Zabara Foundation amongst others, including individual entrepreneurs.
Generation Lab raised an $11M seed round led by Accel.The longevity-focused biotech’s proprietary product, SystemAge™, quantifies biological age via examining the health of an impressive 19 organ systems and functions (including cardiovascular, reproductive, immune, fibrotic, metabolic, and regenerative) as generated from primary blood samples against ‘the physiologic non-linear curve of DNA methylation aging’. Only ten-months post-launch, Generation Lab has seen wildfire adoption by 275 clinics whilst accumulating 300M human-aging data points and achieving 99.9% diagnostic accuracy. The tech boasts pre-symptomatic prediction of many disorders by determining biological dysregulation as confirmed by subsequent clinical tests that may likely not be offered to patients at such early disease onset stages. This offers significant potential for preventative health as well as earlier clinical intervention leading to more positive patient outcomes. Additional investors include: Samsung Next, Zone2, Aoki Labs, Build Your Legacy (BYL) Ventures, and Markham Valley Ventures.
In case you missed it
Origin Bio launches Axis: the first AI model that generates regulatory DNA elements and predicts their function.
Anthrogen introduces Odyssey, the world’s largest and most powerful protein language model.
What we listened to
What we liked on socials channels
Events
Don’t miss it: Last chance to sign up today! Sign up here.
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.












This article comes at such a perfect time. What if C2S-Scale's 'sentences' really do let us program biology, not just predict it, for truly personalized terapies? Makes you wonder about the possibilities.