BioByte 131: μProtein model accelerates protein engineering, epigenetic memory is not just a binary, a multi-component nanoparticle offers new RA treatment, and ABCs for improved cancer therapies
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
Study finds cell memory can be more like a dimmer dial than an on/off switch [Jennifer Chu, MIT News, September 2025]
Why it matters: For decades epigenetic memory has been framed as a binary lock—genes are either ‘on’ or ‘off’—but Palacios et al. illustrate a different picture: gene expression can be stably set to intermediate levels, creating persistent, analog epigenetic memory. This forces a reappraisal of how cellular identity and long-term gene regulation are encoded: the team demonstrates a straightforward molecular substrate for tuning cell-state endpoints rather than only flipping cell fate decisions. Ultimately, this has implications for how we consider developmental biology, disease states, and the design space for programming cellular behaviors.
Palacios et al. use targeted chromatin editing to demonstrate that transiently recruiting DNA- and histone-modifying effectors to promoters produces graded methylation states that map monotonically to steady-state gene expression levels. Crucially, these states remain stable even after the editors are removed, substantiating the idea of analog memory.
Methodologically, the study is perturbative and cross-validated. The team built modular recruitment systems, fusing DNA-targeting proteins such as dCas9, PhlF, and rTetR to writers and erasers such as DNMT3A (a DNA methyltransferase), TET1 (a DNA demethylase), and KRAB (a histone methylation recruiter). After the application of these effectors to multiple loci upstream of a reporter gene and their subsequent withdrawal, they track promoter CpG methylation, histone marks, and transcript levels over time. They arrive at a core empirical result that is simple and consistent: the mean fraction of methylated CpGs in a promoter (the ‘methylation grade’) is set by the pulse of DNMT activity (effector dosage), inversely predicts transcription level, and is temporally robust; in contrast, H3K9me3 (a repressive chromatin mark) decays unless reinforced by DNA methylation. Coupling the experiments with a compact mathematical model, the authors contend that DNA methylation’s capacity for analog memory arises since it can be written and maintained at fractional levels and lacks the same bistable positive-feedback loop architecture that makes certain histone systems behave like switches.
While the results are encouraging, multiple caveats persist. Given that the demonstrations occur in engineered cell lines and targeted loci, factors such as locus-dependence, chromatin context, organismal development, and multi-generational stability remain unresolved. Broader investigations, encompassing broader tissue and in-vivo testing, single-cell lineage assays, and analyses to map interactions between graded methylation and 3D chromatin architecture, are likely necessary to fully parameterize this phenomenon.
Practically, the discovery lays a foundation for future basic and applied epigenetics. It creates opportunities in synthetic biology for constructing circuits and networks requiring tunable expression or programmable analog memory. Moreover, it generates new areas of hypotheses for diseases characterized by perturbed methylation patterns, such as potential connections to lineage infidelity and tumor heterogeneity in cancer.
Papers
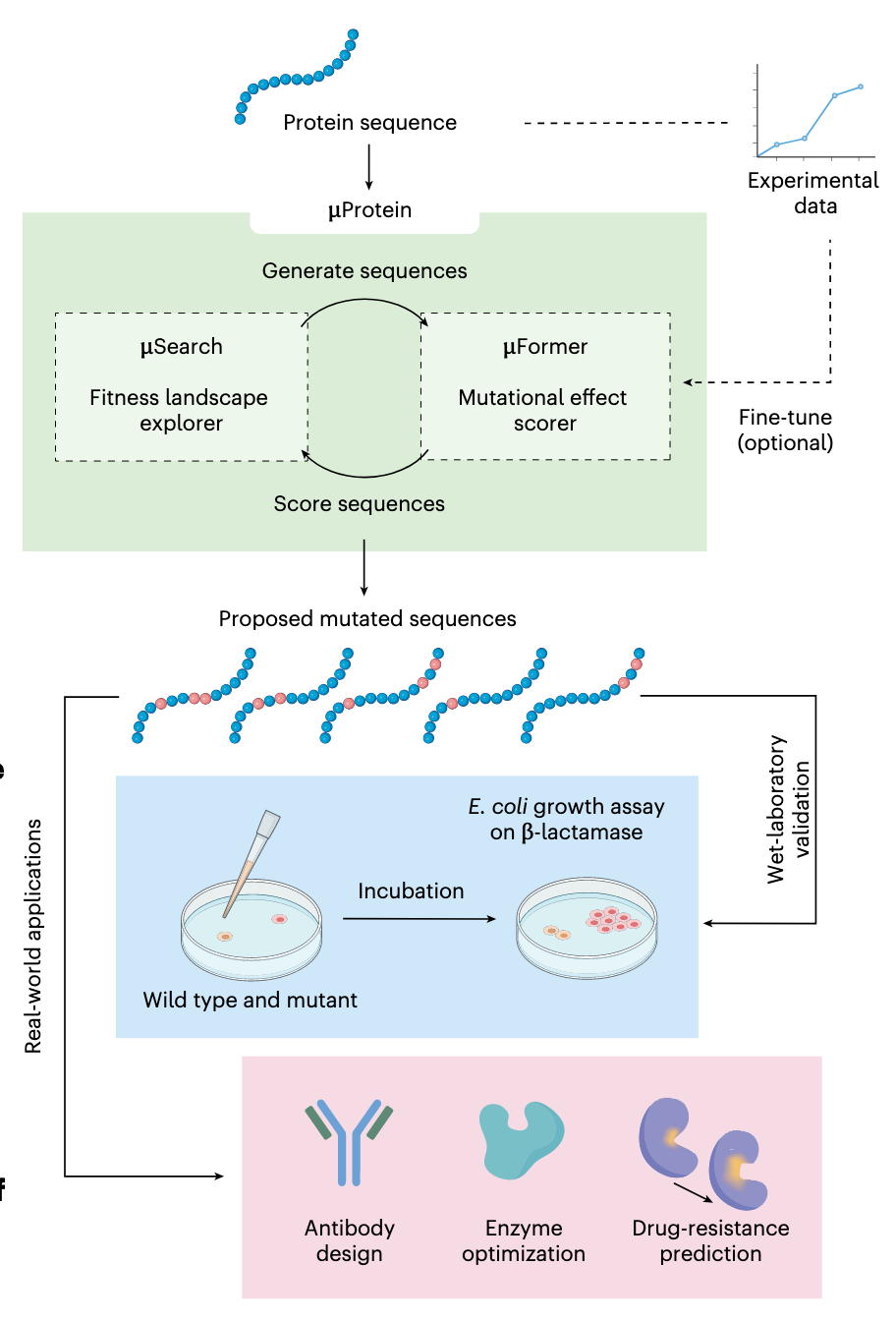
Accelerating protein engineering with fitness landscape modelling and reinforcement learning [Sun et al., Nature Machine Intelligence, September 2025]
Why it matters: Mapping protein sequence to function is the foundational task to protein engineering. This fitness landscape, however, is complex due to the interactions between amino acids in protein sequence. By systematically probing single amino acid substitutions at high throughput, the field has advanced in its ability to measure fitness effects. These tools, however, face the challenge of an exponentially growing combinatorial sequence space with number of mutations, meaning most combinations can’t be tested experimentally. They also face the fact that they need to be tested by a measurable outcome (e.g. cell growth), which cannot be easily carried out for many protein functions. The model proposed here, μProtein, was designed to generalize from limited experimental observations to explore a larger fitness landscape.
μProtein, composed of two models μFormer and μSearch, accelerates protein engineering by combining mutational effect prediction with efficient navigation of the protein fitness landscape.
μFormer is a deep learning fitness prediction model built by pre-training a Pairwise Masked Language Model (PMLM) on more than 30M protein sequences to learn residue-residue dependencies and by adding three scoring models at sequence-, motif- and residue-level, whose outputs are summed to produce a fitness score for any sequence. The model can generalize from limited single-mutant data. μFormer was benchmarked against 16 alternative approaches using ProteinGym, where it performed the best overall performance. It also captures epistasis and supports indel effect prediction.
μSearch uses a Markov decision process to navigate the fitness landscape. In this process, each state is the current sequence and each action is a single amino-acid change. It uses Proximal Policy Optimization (PPO) with exploration noise—to avoid local optima—and two policy networks: one to pick the mutation site and one to pick the amino acid type, using the μFormer score as the reward. The team benchmarked μSearch against 8 exploration algorithms using the open-source FLEXS toolbox. μSearch demonstrated superior sample efficiency on several protein landscapes.
The team tested μProtein’s effectiveness using the TEM-1-cefotaxime system. μFormer was trained on TEM-1 β-lactamase enzyme single-mutant data and μSearch explored millions of possible variants with 2-3 amino acid changes to identify enhanced activity against cefotaxime. The superior identified variants were validated using E. coli growth assays, in which 23.5% of the predicted variants had superior enzymatic activity against cefotaxime versus only 12% of those randomly generated had superior enzymatic activity.
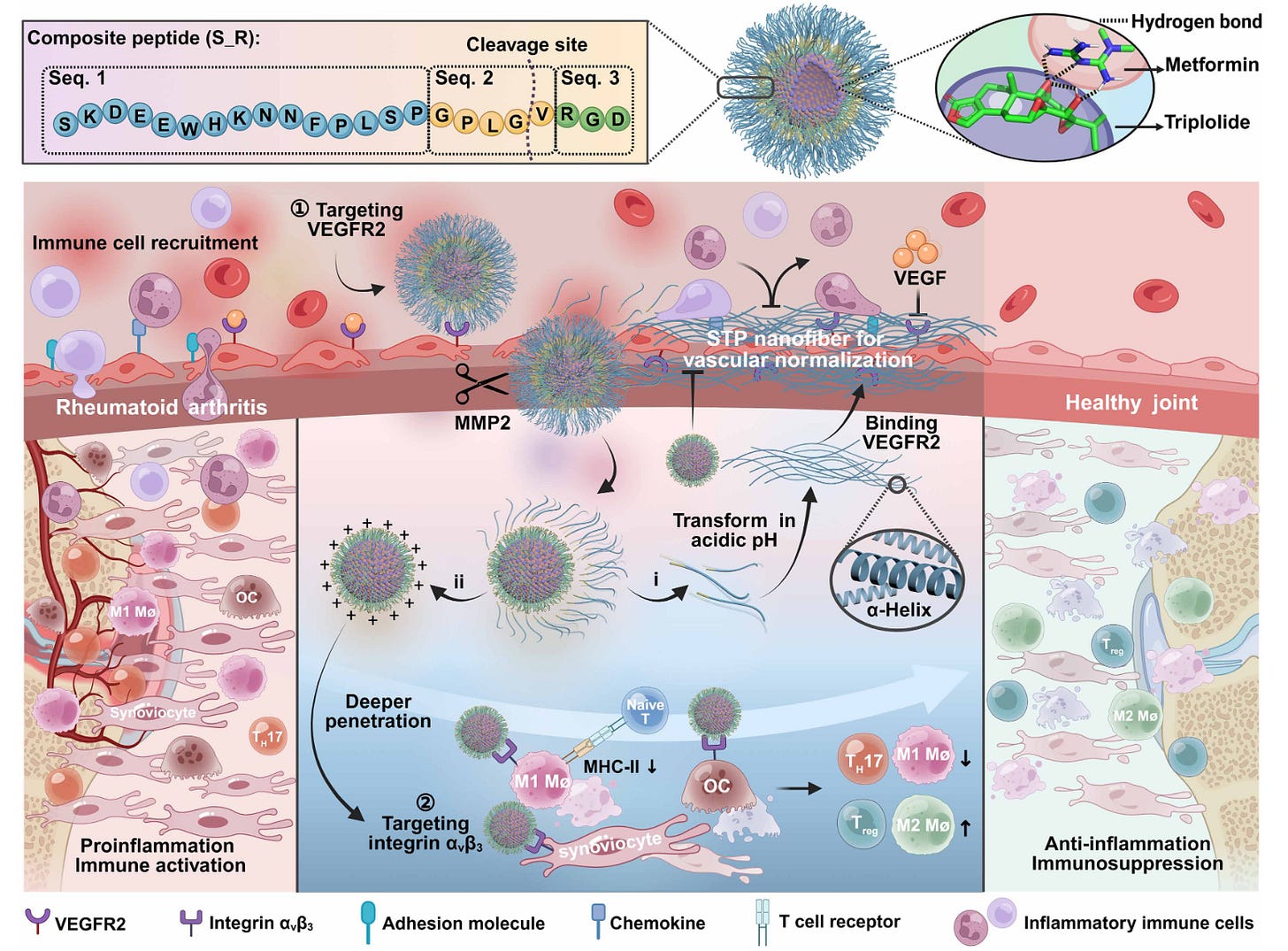
Microenvironment-driven transformable self-assembly nanoplatform enables spatiotemporal remodeling for rheumatoid arthritis therapy [Hu et al., Science Advances, September 2025]
Why it matters: Rheumatoid arthritis (RA) is a chronic autoimmune disease damaging the joints. Standard treatments such as anti-inflammatories (NSAIDs, steroids) blunt the symptoms by curbing some immune stimulation, but don’t correct for vascular leakiness or deeper immune dysregulation. Hu et al. develop a multi-component nanoparticle that alleviates leaky vasculature and immune stimulation, offering a new advanced strategy to treat RA.
Their system is relatively complex, so first let’s illustrate it in some broad strokes. There are two main components of RA that the authors address. The first component is that the vasculature of the joints under RA is leaky—allowing for drugs to easily enter and escape. It also enables immune cells to enter the joint. The second component is that in the joint, immune cells are activated by local cells expressing MHC-II and costimulatory factors such as CD54, CD80, and CD86. The authors aim to, in the local context of the RA, stitch up the leaky vasculature and reduce immune activation.
To address the leaky vasculature, the authors take advantage of a peptide with sequence SKDEEWHKNNFPLSP. This peptide, under acidic conditions (like in RA!) will self-assemble into nanofibers and specifically target VEGFR2 which are expressed on inflamed endothelium. They show that this “blanket” reduces inflammatory cell adhesion while also inhibiting angiogenesis.
To address the immune activation, they combine the functional effects of metformin and triptolide. Metformin has been shown to reduce MHC-II and CD54/CD80/CD86 expression. When cells in the body express this, they can recruit and activate immune cells, which leads in this context to an autoimmune response against the body’s own cells: metformin helps reduce the local native cells from stimulating the autoimmune response. Triptolide is cytotoxic to pro-inflammatory M1 macrophages, osteoclasts, and hyperproliferative synoviocytes, helping to curb the autoimmune response and bone degradation. Metformin and triptolide organically form nanoparticles when mixed, due to metformin’s abundance of nitrogens and triptolide’s numerous oxygens.
To combine the two, they tether the composite peptide (STP–GPLG–VRGD, or S_R) to the triptolide+metformin nanoparticle core using a PEG–lipid anchor. This makes a single unit where the drugs are inside and the peptide sits on the surface. The clever part is how it responds to the RA microenvironment: in inflamed joints, there’s an abundance of MMP2, an enzyme secreted by inflammatory cells that degrades the extracellular matrix. The GPLG linker in S_R is specifically cut by MMP2. That cleavage event splits the construct into two pieces. This setup gives you a double hit: the nanofibers act locally to seal vessels and curb new angiogenesis, while the nanoparticles deliver metformin and triptolide directly to disease-relevant cells, cutting antigen presentation and killing hyperinflammatory or bone-destructive cells. Joints treated with the nanoparticle system showed over sixfold higher drug retention and significantly reduced inflammation, with restored immune balance (i.e. more Tregs, fewer TH17 cells) compared to free drug or control treatments.
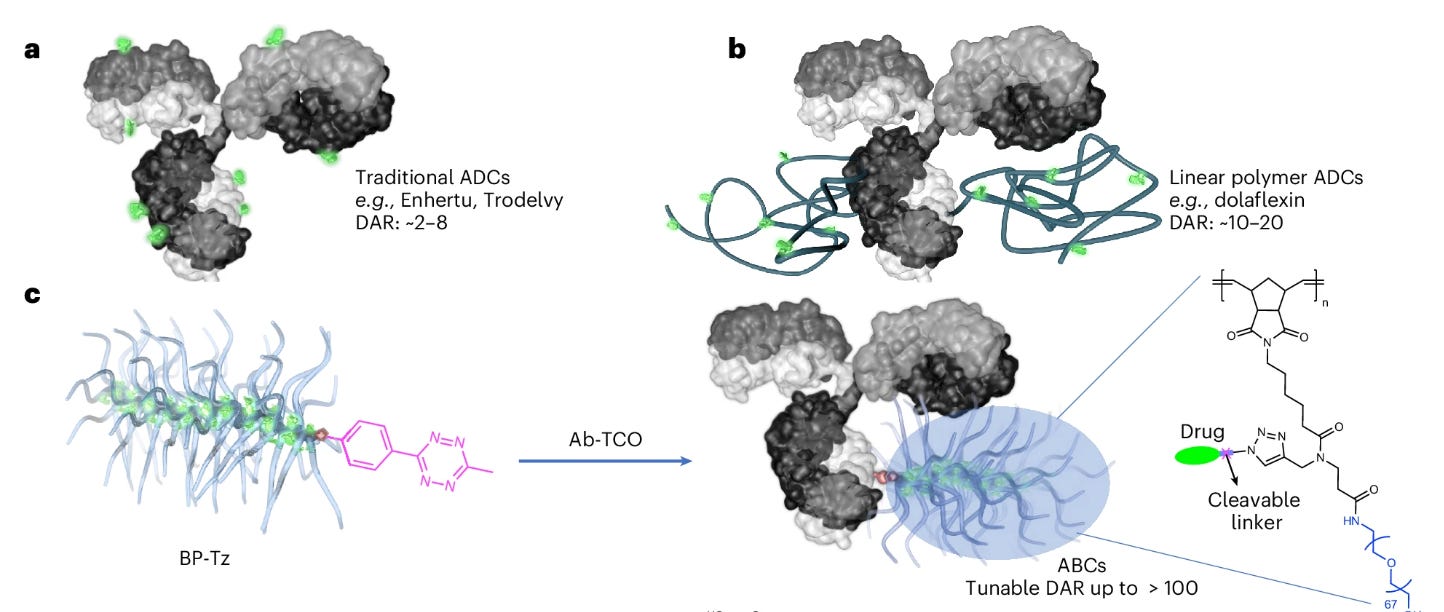
Antibody–bottlebrush prodrug conjugates for targeted cancer therapy [Liu et al., Nature Biotechnology, September 2025]
Why it matters: One promising avenue of development in the targeted therapy space is antibody-drug conjugates (ADCs). ADCs consist of small molecules linked to a monoclonal antibody which helps deliver drugs to targets of interest. However, current ADCs are restricted in the number of drug molecules they can carry per antibody (drug-antibody ratio or DAR), meaning the platform is limited to agents with high cytotoxicity risks with few mechanisms of action. In this paper, the authors describe their antibody-bottlebrush prodrug conjugates (ABCs) technology which features the ability to modulate DARs, opening the door for the delivery of less toxic drugs with a broader range of mechanisms. Such improvements should ideally pave the way for more safe and efficacious therapies for a variety of cancers.
ADCs have limited payload capacity with a single antibody usually carrying no more than 8 drug molecules to preserve desirable properties like stability and solubility. Previous approaches to increase DARs used polymers or nanoparticle encapsulation to increase potential payloads; however, these methods also exposed the payloads to the external environment, meaning that the overall properties of the construct vary significantly based on the drug being used. To obtain a more predictable alternative, the authors developed ABCs which link antibodies with BPDs. BPDs are branched polymers where each subunit has two side chains—one polyethylene glycol (PEG) side chain and one drug molecule attached with a cleavable linker. The long hydrophilic PEG side chains “shied” the linkers and drug molecules from the external environment and confer much more stable properties to the overall construct. The repeating brush-like structure of BPDs makes the associated construct DAR highly modulatable and allows for precise tuning of therapeutic dosage. The authors also use click chemistry reactions to facilitate the conjugation between the targeting antibody and larger BPD structure.
To validate ABCs, the team studied the properties of BPDs combined with trastuzumab, a HER2-targeting antibody. Apart from studies of basic synthesizability and in vitro uptake, additional experiments tested ABC efficacy when using conventional drug payloads as well as candidates with mechanisms of action that have not yet been proven in clinical settings. Interestingly, certain ABCs also showed higher effect in mouse models when compared with existing clinical ADC options. The authors concluded that ABCs can achieve drug-antibody ratios up to two orders of magnitude higher than current ADC alternatives. Noting that high DAR values can limit the effectiveness of ADCs, ABCs are far more suitable platforms for less potent drugs and may dramatically expand the repertoire of candidates that can be integrated into targeted therapy approaches. While more clinically relevant features of ABCs like biodistribution and pharmacokinetics are unknown, the new approach is a hopefully promising step towards improved targeted therapies for a variety of cancers. It will be interesting to see how further optimization of ABC design with other polymer chains and linkers may improve the platform as a whole.
Notable deals
Enveda announced the close of an oversubscribed $150M Series D led by Premji Invest, achieving unicorn status. This financing round was preceded by a $150M Series C earlier this year and was accompanied by news of the appointment of Dr. Mikael Dolsten—former CSO and President of Worldwide R&D at Pfizer—to Enveda’s Board of Directors. The company also elected to announce their first patient enrollment in a Phase 1b clinical trial for ENV-294, the company’s lead program intended to treat atopic dermatitis. A mere five years out from seed financing, one investor quoted in the briefing remarked on the “unprecedented speed, scale, and efficiency” of Enveda’s development of small molecule therapeutics, in large part due to the combination of the company’s exploration of existing chemical diversity in nature with AI and their high-throughput lab automation for the multitudinous analysis. In addition to ENV-294, Enveda’s pipeline hosts 15 other preclinical programs, including four IND-enabling programs and over a dozen more development candidates. The funds from this round will go towards advancing several clinical programs through Phase 1b and Phase 2 trials as well as support new upcoming IND filings for therapeutic candidates intended to treat various other indications such as asthma, inflammatory bowel disease, and obesity. Other investors in the round include Baillie Gifford, Kinnevik, Lingotto Investment Management, Peakline Partners, FPV, Socium Ventures, Dimension, Level Ventures, Henry Kravis, IA Ventures, and Lux Capital.
Ridge Biotechnologies has emerged from stealth with an oversubscribed $25M seed round led by Sutter Hill Ventures (which also incubated the company). Leveraging their wet lab expertise centered on cell-free synthesis and screening, Ridge Bio has built proprietary ML models which they are using to surmount enzyme-related biological challenges at unprecedented speeds. The potential applications of their platform span not only the improvement and design of a wide range of therapeutic modalities—including ADCs, CAR-T therapies, and radiotherapies—but also the design of better prodrugs, biocatalysts, and drug stability enhancers. NativeLink and ProTrigger, their current two leading products, are demonstrative of the power of their platform, enabling significant modification of proteins and peptides without significant genetic alterations and triggered release of payloads in target tissue to reduce off-target effects and toxicity respectively.
Odyssey Therapeutics secured $213M in a Series D financing round. The funding will be utilized to carry out several proof of concept studies for the company’s leading programs in autoimmune and inflammation indications, strengthening Odyssey’s preclinical pipeline. Odyssey had previously planned to go public this summer, but abandoned the efforts amidst the market turbulence and ongoing difficult environment for biotech. Nevertheless, founder Gary Glick has his sights set on the long-term, professing in the Fierce Biotech deal coverage the desire to see many of the company’s assets through commercialization. This round appears to pave the way to this goal, giving the company three years of runway and confidence at present of securing the next round. All existing investors participated in the round, with new additions of Affinity Asset Advisors, Dimension Capital, Jeito Capital, Lightspeed Ventures, TPG Life Sciences Innovations, and Wedbush Healthcare Partners.
NRG Therapeutics closes £50m Series B round to advance proof of concept studies for leading ALS/motor neuron disease drug program.The round will also enable meaningful clinical data generation in patients with Parkinson’s Disease (PD) through Phase 1b. NRG’s lead candidate, NRG5051, is part of a novel class of small molecule mitochondrial permeability transition pore (mPTP) inhibitors designed for successful brain penetration via oral delivery route. In preclinical models, the drug conveys protective effects against mitochondrial degradation in the brain, an identified underlying pathology commonly seen in PD and similar neurodegenerative disorders. The Series B was led by SV Health Investors’ Dementia Discovery Fund (DDF) with participation from British Business Bank, M Ventures, Novartis Venture Fund, Criteria Bio Ventures, Omega Funds, Brandon Capital, and founding investor, Parkinson’s UK, via their drug discovery arm, Parkinson’s Virtual Biotech.
In case you missed it
First-Ever FDA Acceptance to Waive Clinical Efficacy Studies for Monoclonal Antibody Biosimilars
What we listened to
What we liked on socials channels
Events
Evolved Technology is running the third annual AI x Bio Hackathon from November 7-16, and in-person regional hackathons in SF and Boston in October. Winning teams will be invited to NVIDIA headquarters to showcase their work.
Learn more here.
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.