BioByte 130: Understanding Inner Speech for Neuroprostheses, Phylogenetic Relationships Hidden within Evo 2, and Nanosyringes to Deliver Diverse Biomolecules
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
Study of promising speech-enabling interface offers hope for restoring communication [Bruce Goldman, Stanford Medicine, August 2025]
A new publication from the Neural Prosthetics Translational Lab at Stanford Medicine explores a key question: is inner speech represented in the motor cortex, and can it be decoded quickly and safely enough for speech-restoring neuroprostheses? Although previous work from the lab has demonstrated that brain-computer interfaces (BCIs) can reliably extract and translate signals from paralyzed individuals attempting to speak or write, whether inner speech - i.e. imagining the sounds or feelings of speaking - can be similarly used is an open area of inquiry. Unlocking inner speech as a modality could enable more natural, fluent communication, while reducing the fatigue and articulatory effort often required of users.
To investigate this, Kunz et al. recorded data from microelectrode arrays implanted in four individuals with severe paralysis. They reveal that inner speech produces a robust, speech-like neural geometry in the ventral and mid-precentral cortex. Notably, inner speech is not a degraded, noisy shadow of attempted speech, but instead forms a scaled, separable representation - the authors identify a ‘motor-intent’ neural dimension distinguishing inner from attempted speech, providing a distinct substrate for decoding.
The decoding pipeline the authors deploy is straightforward: neural features are fed into a recurrent neural network that generates phoneme (atomic building blocks of speech) probabilities every ~80ms, these logits are then converted into words and sentences using a language model, and a text-to-speech engine renders audio. In closed-loop tests involving explicit cues, the system decoded imagined sentences in real-time using a 125,000 word vocabulary and achieved encouraging accuracies - ranging from 46 - 74%.
The paper concludes by addressing a crucial ethical issue: is freeform, uninstructed inner speech decodable? To probe this, participants were prompted with open-ended tasks such as sequence recall, counting, and word associations. The corresponding inner speech signals were, in principle, interpretable - though predictive accuracy varied significantly across participants and tasks. Critically, the team also demonstrated two separate, high-accuracy safety mechanisms to prevent inadvertent translation of inner speech. The first control involves labeling attempted speech patterns with their appropriate phonemes and labeling inner speech patterns with a ‘silence’ token, effectively suppressing the latter from the output. The second control involves the user needing to unlock the system’s ability to perform inner speech decoding with an inner speech keyword (e.g. a mental password).
Despite limitations in sample size, decoding variability, and hardware constraints, the study offers an exciting proof-of-principle for leveraging inner speech in BCIs in a privacy-preserving way.
Finding the Tree of Life in Evo 2 [Pearce et al., Goodfire, August 2025]
Foundation models have demonstrated an ability to internalize complex structures in their embeddings - for example, language models encode syntactic and semantic structures as geometric patterns in high-dimensional space. A recent publication by Pearce et al. finds that Evo 2 - a 40B parameter, long-context DNA foundation model trained on 8.8 trillion nucleotides from sequences spanning bacteria, archaea, eukaryotes, and bacteriophages - implicitly encodes species identity and evolutionary relationships as a structured, geometric object: a curved phylogenetic manifold. Although this may seem esoteric, discovering and interpreting these structures in scientific models enables us to extract the reasoning behind model predictions, repurpose the model’s internal representations for novel tasks, and unlock a human-interpretable way to prompt, steer, and edit the model.
To uncover this phylogenetic structure, the authors analyzed 2400 bacterial species from the Genome Taxonomy Database by randomly sampling 4kbp genomic windows per species and averaging Evo 2’s internal activations - creating species-level embeddings. While projecting and visualizing these species vectors revealed taxonomy-aware clustering, the key insight emerged from the geometry of the feature manifold: they built a k-nearest neighbors graph (ensuring a connected graph) and computed geodesic distances along the resulting manifold. These geodesics were linearly correlated with the true phylogenetic branch-length distances (excluding a small set of outliers representing the order Mycoplasmatales), demonstrating that the geometry of the model’s embeddings encodes evolutionary history in an interpretable manner.
Having discovered that phylogenetic relationships form a curved manifold, the final question the team addresses is whether a simpler structure underlies this complexity. In other words, this can be broken down into two finer, intertwined points: (a) can evolutionary organization be represented by a clean, ‘flat’ coordinate map? (b) can the curvature be explained by secondary genomic features warping and modulating this map? By training a compact encoder to jointly reconstruct embeddings and predict evolutionary distances, they discovered a low-dimensional subspace that captures most of the variance underpinning phylogenetic organization in Evo 2’s space and exhibits high correlation (0.89+) between the subspace distance metric and phylogenetic distance. Moreover, the authors show that the learned subspace correlates with genomic statistics (ex. codon frequency) and a decision tree using genome-wide statistics of up to 4-mers can predict subspace activations. Ultimately, this substantiates the idea that the manifold is not an abstract mathematical artifact, but one grounded in meaningful biological signals.
The authors acknowledge that these are intermediate results—verifying the robustness of the approach across different domains of life and sets of parameters is a critical next step. Nonetheless, the findings lay the groundwork for unearthing these “latent ontologies”, which could eventually open up applications from accelerating functional annotation of genes and metagenomic sequences to building a predictive engine for complex phenotypes.
Papers
Targeted delivery of diverse biomolecules with engineered bacterial nanosyringes [Kreitz et al., Nature Biotechnology, August 2025]
Why it matters: Bacteria and their viruses use spring-loaded “nanosyringes” to inject proteins into nearby cells. Kreitz et al. turn one of these machines, the Photorhabdus virulence cassette (PVC), into a modular delivery platform called SPEAR that can deliver not just proteins but also Cas9 ribo nucleoproteins (RNPs) and single-stranded DNA (ssDNA), and can be retargeted to chosen cell types. This effort expands the biologics delivery toolbox, enabling delivery of diverse biomolecules into mammalian cells without relying on cellular import mechanisms.
Bacteria and their viruses use spring-loaded “nanosyringes” to inject proteins into nearby cells. Kreitz et al. turn one of these machines—the Photorhabdus virulence cassette (PVC)—into a modular delivery platform called SPEAR that can deliver not just proteins but also Cas9 ribonucleoproteins (RNPs) and single-stranded DNA (ssDNA), and can be retargeted to chosen cell types. This effort expands the biologics delivery toolbox, enabling delivery of diverse biomolecules into mammalian cells without relying on cellular import mechanisms.
Think of a PVC as a spring-loaded nanosyringe: a tail fiber latches onto a receptor, the sheath snaps shut, and a spike punches through the membrane to drop off cargo. Older PVC work stuffed proteins down the hollow tube, which tends to unfold big cargos and can’t carry guide RNA with Cas9. Kreitz et al. flip the loading site to the spike itself: they fuse cargos to the spike base (Pvc8) or tip (Pvc10), so cargos stay folded. When they co-express Pvc8/Pvc10–Cas9 with an sgRNA, the assembled particles purify with about a five-log enrichment of sgRNA and edit human cells without any transfection—the RNP rides the syringe in a ready-to-cut state. The targeting arm (Pvc13) is modular too: dropping in antibody-friendly tags (SpyTag or SNAP) lets them “click” on scFvs or full mAbs, so the same syringe can be pointed at different cell types with simple conjugation swaps.
Originally PVC was just used to deliver protein—Kreitz et al. use creative methods to expand this toolbox to RNPs, ssDNA, and even split-protein systems. For RNPs, they shift loading to the spike—fusing Cas9 to Pvc8/Pvc10—so Cas9 stays folded and co-purifies with its sgRNA, producing editing-competent particles that cut human cells without any transfection. For ssDNA HDR, they add an HUH endonuclease onto Pvc10 to covalently tether a single-stranded oligo (ssODN) to the spike, enabling co-delivery of Cas9 RNP + ssODN and precise 6-bp insertions. For the zinc-finger deaminase (ZFD) split editor, they combine spike-loading with classic tube-loading to package both halves in one PVC, reconstituting base-editing activity at the target and pushing SPEAR beyond single-polypeptide cargos.
The power of this system is its modularity. They deliver diverse payloads into multiple mammalian cell types by engineering the spike, the tail fiber, and the conjugation chemistry (SpyTag/SpyCatcher, SNAP, and HUH-based ssDNA tethering). In short, Kreitz et al. deliver a bacterially manufacturable, antibody-swappable injector that delivers diverse payloads including RNPs and ssDNA directly into cells without transfection or endocytotic pathways, enabling targeting of challenging targets such as senescent cells.
Generative Medical Event Models Improve with Scale [Waxler et al., arXiv, August 2025]
Why it matters: Guiding a patient towards optimal health outcomes requires understanding of their longitudinal medical history, addressing outcome uncertainty and their goals and values. Foundation models trained on Real World Data (RWD) to generate Real World Evidence (RWE) is a scalable way of turning raw data into actionable, personalized insights at the point of care.
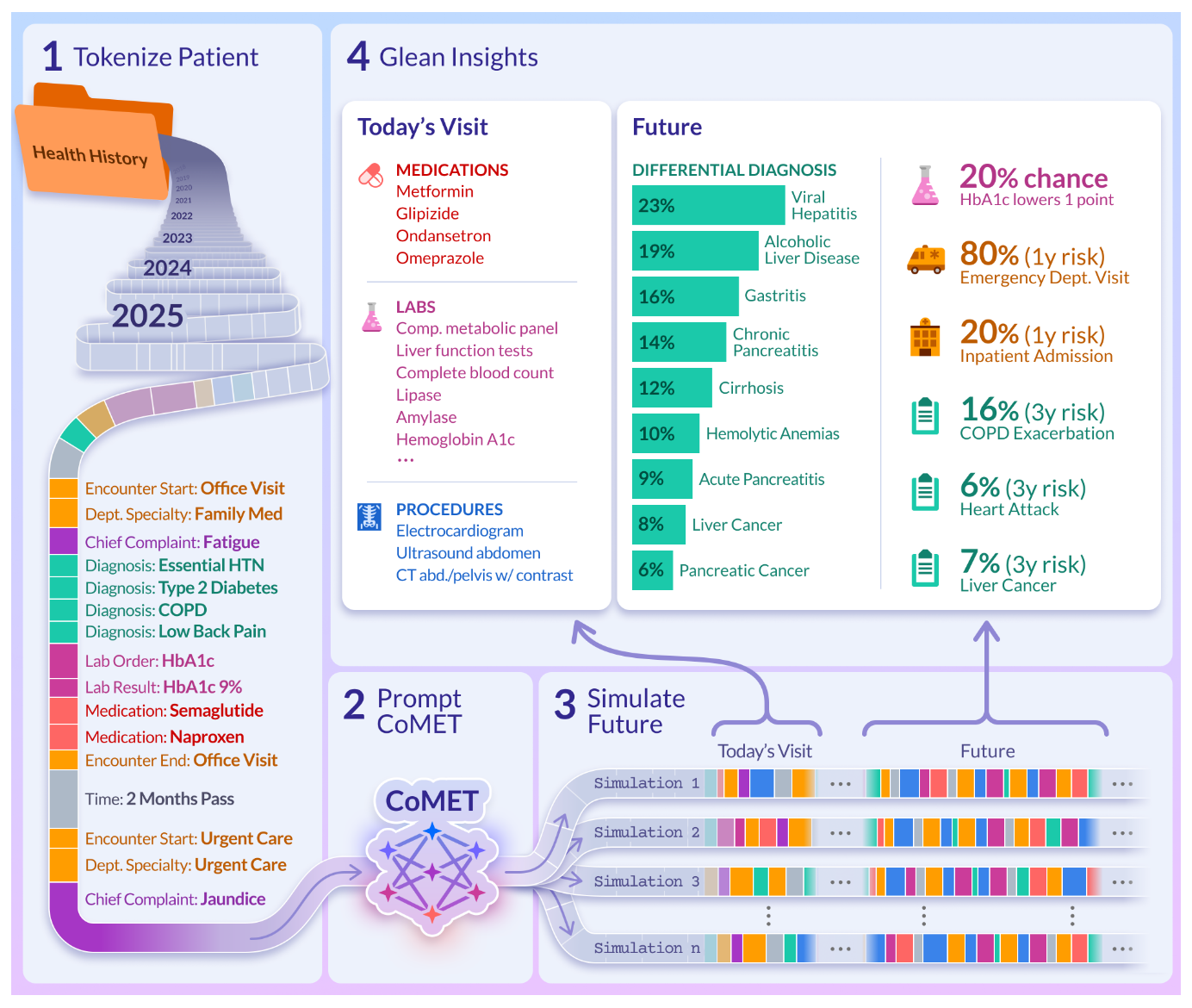
Researchers from Epic Systems, Microsoft Research and Yale introduce Cosmos Medical Event Transformer (CoMET) models; a family of decoder transformer models pre-trained on 118M patients representing 115B discrete medical events. CoMET was trained on the Epic Cosmos dataset: a collection of records for more than 300M patients and 16.3B encounters, de-duplicated and combined into a single, integrated longitudinal record for each patient. The platform unifies a variety of data including lab results, diagnoses, medications and procedures as well as social drivers of health, cancer, genomic variants and patient-reported outcomes. CoMET learns by predicting the next medical event in a patient’s journey. At inference time, the model is prompted with the patient’s medical history and simulates trajectories by generating the next events.
Foundation models pre-trained on RWD from patient journeys have shown promise in their ability to quantitatively predict the likelihood of events over specific time intervals, by learning latent representations of complete patient records. However, due to the size of available datasets, previous studies have not systematically investigated the impact of model size and computation on the outcomes of these models. According to the team, this is the largest scaling-law study on real-world patient journeys.
The family of CoMET models (small, medium and large) demonstrated:
Strong predictive performance in:
Medical event sequences: On 30-day readmission prediction, CoMET-L reached an AUCROC of 0.770, compared to 0.717 for the best supervised model. On multiple outcome tasks (T2DM complications, chronic kidney disease, emergency procedures), performance improved smoothly as training loss decreased.
Personalized future disease risk: On incident disease prediction (six diseases across a two-year horizon with a prevalence c.1.5%), CoMET-L achieved higher PR-AUC than supervised models on all six tasks.
Early, quantitative differential diagnoses: For hepatopancreatobiliary (HPB) diseases and rheumatic diseases, CoMET-L correctly flagged more than 50% of patients with their eventual diagnosis by the final prediction time, and more than 25% of patients weeks ahead of diagnosis.
Patient’s interactions with the health system
Scaling up model and dataset size predictability decreases training loss and that improves downstream evaluation scores
The Therapeutic Nanobody Profiler: characterising and predicting nanobody developability to improve therapeutic design [Gordon et al., bioRxiv, August 2025]
Why it matters: When designing an effective drug, it is important that the therapeutic not only has favorable binding characteristics but also demonstrates suitable developability characteristics (stability, solubility, aggregation, etc.). Measuring these properties can involve expensive physical validation, meaning that reliable in silico predictors would drastically reduce the time and effort required for such drug development tasks. In this paper, Gordon et al. build previous work that measures the developability of antibodies to develop a similar framework for nanobodies. This work paves the way for tools to further support the development of a burgeoning class of therapeutics.
While antibodies are the largest class of biologics, the emergence of new therapeutic modalities like nanobodies require new bespoke developability measurement tools to aid in their overall design. Previously, a myriad of laborious experimental techniques have been used to measure crucial properties like aggregation, non-specific binding, melting temperatures, and other formulation-influencing factors. In the case of antibodies, computational efforts like the Therapeutic Antibody Profiler (TAP) use groups of in silico structural and chemical features to predict developability. However, the structural dissimilarities between nanobodies and antibodies require a new set of features to determine the former’s developability. In this paper, the authors present the Therapeutic Nanobody Profiler (TNP) that is calibrated on the features of 36 clinical-stage nanobodies. Furthermore, the paper also investigates the concurrence between TNP results and experimentally derived characteristics of an additional 72 nanobodies.
The team first investigated how many of the original TAP features for antibodies could be carried over to nanobody profiling. Focusing mainly on CDR3 loop features, it was found that nanobody developability is not as constrained as antibodies, with the clinical-stage candidates showing a wide distribution of lengths. Additionally, the authors also assessed how CDR3 loop conformations potentially affected developability - the measured candidates formed two distinct classes but neither class seemed to confer a higher level of developability than the other. On top of CDR3 characteristics, the team chose to include surface level features like patches of surface hydrophobicity, positive charge and negative charge across the CDR vicinity, bringing TNP to encompass a total of six features. The authors did note that this set of six features could also be exchanged for other characteristics, but the current set was chosen for its relative interpretability. Additionally, it was found that TNP could return “red flag” classifications for certain features of clinical-stage targets, pointing to a need for further calibration. Nonetheless, it will be interesting to see how profiling tools may be developed for a wider range of therapeutics in the future, and how TAP and TNP-style software can potentially accelerate drug development campaigns.
Notable deals
With payments of up to $1.2B, AbbVie agreed to acquire Gilgamesh Pharmaceutical’s lead investigational candidate, Bretisilocin. The drug, a 5-HT2A receptor agonist and 5-HT releaser, is a psychedelic compound, designed to reduce the duration of the psychedelic experience without compromising therapeutic results. Gilgamesh recently announced positive Phase IIa results, demonstrating a statistically significant reduction in MDD symptoms. As a part of the deal, they will also spin off Gilgamesh Pharma Inc., which will house the remainder of the company’s programs and employees.
Emerging from stealth, Arnatar Therapeutics announced they received an Orphan Drug Designation and Rare Pediatric Disease Designation for their Alagille Syndrome candidate, ART4, an antisense oligonucleotide that upregulates expression of JAG1. These two designations will afford the company significant benefits, including seven years of market exclusivity and a Priority Review Voucher upon approval. With their 2024 $52M Series A—led by 3E Bioventures and Eight Roads—Arnatar currently has seven assets in their pipeline, including ART1, an siRNA candidate in early clinical development for blood pressure control.
Leal Therapeutics secured $30M in funding in their Series A, a round led by SV Health Investors’ Dementia Discovery Fund. Existing investors including OrbiMed, Euclidean Capital, and Newpath Partners also participated in the round. The funding will be used to progress two programs, LTX-001 and LTX-002, through a clinical trial in schizophrenia patients and the collection of initial clinical data respectively. LTX-001 is a small molecule inhibitor of glutaminase, with additional potential applications in bipolar disorder, ALS, and major depressive disorder, while LTX-002 is an antisense oligonucleotide for genetic or sporadic ALS.
The CAR-T therapy company Wugen raised $115M to fund the pivotal trial of WU-CART-007, an allogeneic CAR-T therapy for relapsed and refractory leukemia (T-ALL) and lymphoma (T-LBL). In its Phase I/II trials, admission of the drug at the recommended dose resulted in a 73% composite complete remission rate and 91% overall response rate. The money will also fund engagement with the FDA and EMA, as well as manufacturing preparations. Funding was led by Fidelity Management & Research Company with participation from Tybourne Capital Management, Lightchain Capital, and RiverVest Venture Partners among others.
In case you missed it
Accelerating life sciences research
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.