BioByte 129: strategies to drug intrinsically disordered proteins , in silico prediction of RNA-LNP efficacy, and minibinder design to address multidrug-resistant bacterial infections
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
Hitting ‘undruggable’ disease targets [Baker Lab, Institute for Protein Design, July 2025]
Intrinsically disordered proteins and regions (IDPs / IDRs) are central to signaling and disease but have resisted conventional binder design since they lack a stable conformation. Through two complementary papers released by the Baker Lab, researchers reframe IDP plasticity as a design opportunity. The teams used generative protein design to induce and capture binding competent conformations, leading to high-affinity, high-specificity protein binders across in vitro and in vivo (cellular) experiments.
One study - published in Science - introduces the ‘logos’ strategy. This entails developing a library of scaffolds and pocket modules that can be recombined and tailored to match arbitrary disordered sequences. Targets are threaded through these templates, the best matches are refined with ML sequence design, and pockets are tuned with diffusion methods to achieve atomic complementarity. In a one-shot campaign, the authors reported binders for 39 of 43 diverse IDR targets (with typical Kd’s from ~100pM to 100nM, and testing ~22 designs per target) and demonstrated functional readouts, including a dynorphin binder that blocked opioid signaling in cultured human cells. The pipeline emphasizes modularity and combinatorial pocket assembly in order to provide a general solution to sequence-based recognition in IDPs.
The second study - published in Nature - focused on applying RFdiffusion to co-sample target and binder conformations, allowing dynamic molecular shifts during design to reveal favorable induced-fit complexes. Operating solely from sequence, the team generated nanomolar-affinity binders (~3-100 nM) for targets such as amylin, G3BP1, and prion core fragments. Functional efficacy is exhibited again - one such case involving amylin binders dissolving and disassembling amyloid fibrils and rerouting amylin to lysosomes.
Cumulatively, the papers offer a robust proof-of-principle for transforming disorder into a tractable engineering challenge. The performance of this approach under native cellular complexities (e.g. phase separation, post-translational modifications) and therapeutically-relevant constraints - such as developability testing - remains an interesting question for future investigation.
Papers
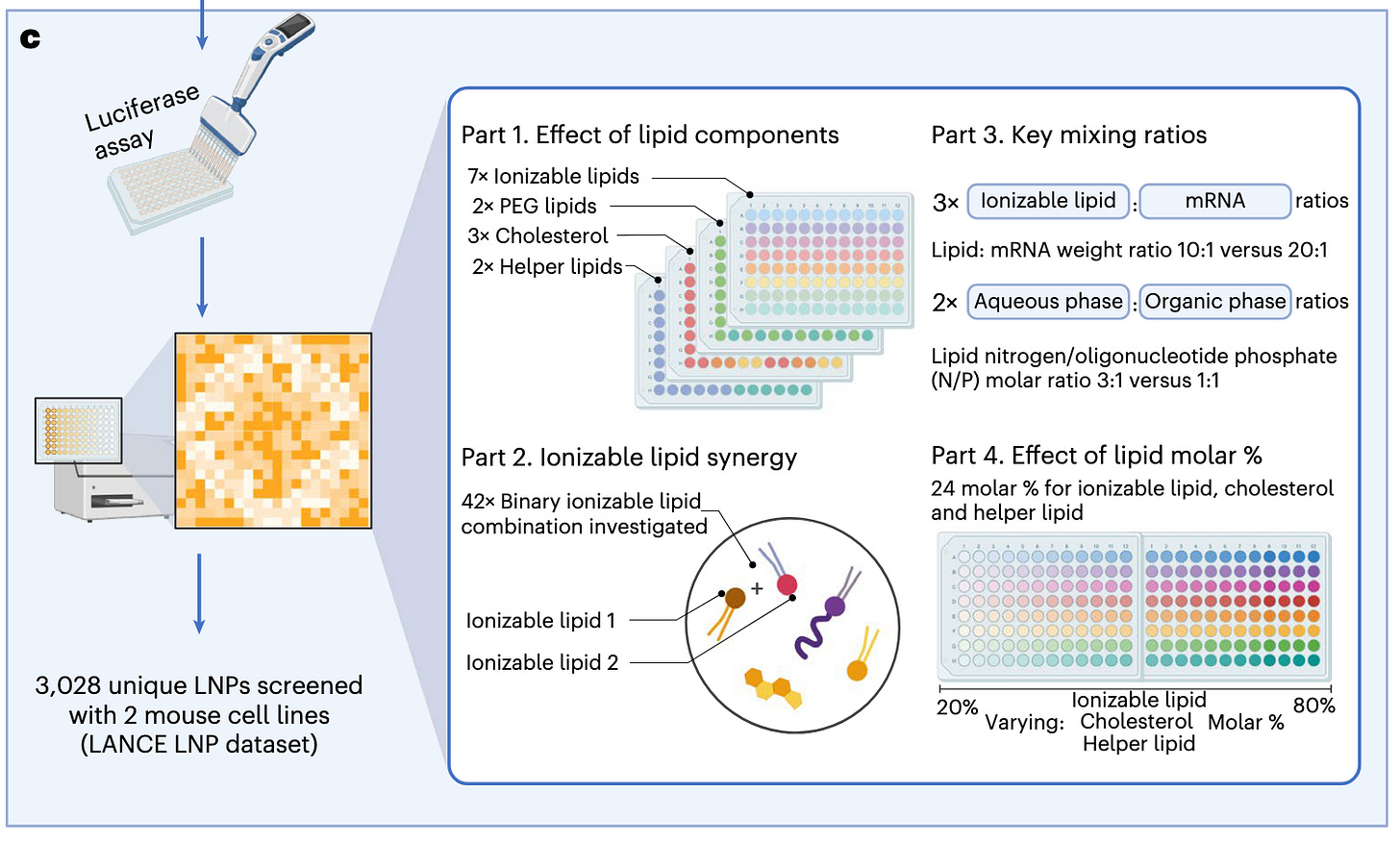
Designing lipid nanoparticles using a transformer-based neural network [Chan et al., Nature Nanotechnology, August 2025]
Why it matters: RNA-LNPs are an increasingly important tool in the therapeutic arsenal. The effectiveness of this modality is determined, in part, by its lipid components and its ratios, and each RNA-LNP needs to be optimized for its application. Exploration of the available individual lipid chemical space, its combinations/ratios and formulation features are not systemically explored, which may contribute to an under optimized drug. The authors of this paper developed a deep learning model which predicts efficacy of an RNA-LNP in silico, which allows wider exploration of lipid combination and formulations leading to potentially more efficacious therapies.
RNA-based lipid nanoparticles (LNPs) are composed of four lipid classes, and their function emerges from the lipid structures and ratios. Each application requires a different composition. Optimizing single lipid molecules or manually selecting features is limiting as it restricts the chemical space available and lacks formulation and composition insights.
The authors of this paper present Composite Material Transformer (COMET), which encodes molecular structures, molar percentages and synthesis parameters in a transformer architecture. The model was trained on the LANCE (Lipid-RNA Nanoparticle Composition and Efficacy) dataset, which includes over 3K LNPs. COMET’s in silico hits from a separate virtual library were experimentally validated. All hits outperformed clinically approved LNPs in DC2.4 and B16-F10 cells.
The team of researchers end with a few caveats: i) COMET’s advantage over random forest depends on data size and complexity, but its transformer architecture will benefit from increasing data sizes from high-throughput experimentation, ii) COMET highlights top from mediocre LNPs well, but optimizing within the high-performing region requires “more discriminative power” and iii) since COMET is trained on in vitro data, many of the hits won't correlate to in vivo efficacy.
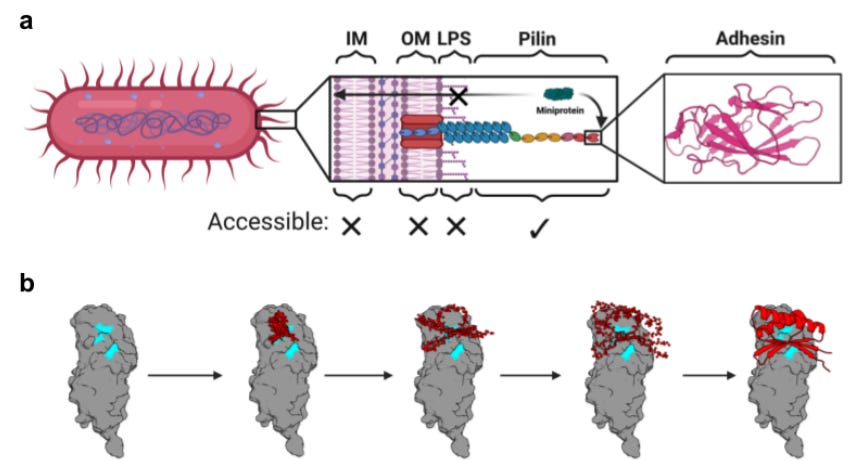
De Novo Design of Miniprotein Inhibitors of Bacterial Adhesins [Chazin-Gray et al., bioRxiv, August 2025]
Why It Matters: De novo protein design capabilities have shown rapid improvement in areas like minibinder generation. However, a binder with high affinity to a given target in vitro does not always have the same activity in vivo. One such mechanism that limits binding is the presence of shielding surface molecules on cell membranes that block access to cell surface targets. Recognizing the limited accessibility of conventional surface antigens, the authors of this paper present a minibinder design approach for bacterial adhesins that lay beyond the reach of shielding surface lipopolysaccharides and discuss the potential for such a strategy to fight multidrug-resistant bacterial infections.
The treatment of multidrug-resistant (MDR) bacteria is an ongoing challenge in modern healthcare, with conditions like urinary tract infections (UTIs) representing nearly 15% of antibiotic prescriptions. Noting the huge gap in timespan between newly approved antibiotics, and their consequent disruption of patients’ microbiomes, a team of scientists have demonstrated a novel method of targeting MDR bacteria using de novo protein minibinders. While generative protein design methods have enabled the rapid development of high-affinity minibinders for nearly any target, in vitro experimental performance often fails to translate to in vivo settings because of surface shielding lipopolysaccharide fibers that occlude surface target antigens. To bypass this, the authors reason that bacterial adhesins, specifically chaperone-usher pathway (CUP) pili adhesins are a suitable alternative target, as they are present on the ends of extracellular bacterial fibers and are responsible for the establishment and persistence of infection. Focusing on both UTIs and catheter-associated UTIs (CAUTIs), the authors develop minibinders for three different classes of adhesins from widespread MDR bacteria.
Beginning with uropathogenic E.coli infections, the authors selected the mannose binding FimH binding adhesin. After characterizing the protein as having two distinct low and high-affinity conformation states (LAS/HAS), they used a standard pipeline of RFdiffusion to generate minibinder backbones followed by sequence generation with ProteinMPNN. Lastly, AlphaFold2 Initial Guess was used to score potential designs, which were also refined by partial diffusion with RFdiffusion again. Crucially, designs were made for both the low and high affinity states of the FimH domain. To experimentally test their designs, the authors used a cDNA display assay to yield 96 enriched minibinders, with the majority of candidates being LAS targets. The overall strongest binder, named F7, was found to have nanomolar affinity, and most interestingly, was capable of biasing the FimH binding domain conformational landscape towards the low affinity state. Finally, F7 was also found to exhibit strong binding to FimH targets on different uropathogenic strains, demonstrating that a single minibinder was capable of handling strains within the same clonal group.
Moving onto A. baumannii bacteria implicated in caUTIs, the authors focused on the Abp1 and Abp2 binding adhesins. Using a nearly identical pipeline as the FimH binders, the authors first characterized the conformational landscape of both targets before using a yeast surface display assay to yield a preliminary batch for further optimization. Further experiments yielded the A7 minibinder with strong affinity to both Abp1 and Abp2 and was capable of disrupting bacterial biofilms in an environment akin to fibrinogen-coated catheters. Finally, both the F7 and A7 binders were validated in mouse models of UTI and caUTI respectively. F7 binders showed nearly a 2 log reduction in bacterial burden, while A7 could only manage a 1 log reduction.
While the RFdiffusion/ProteinMPNN pipeline used in this study has been demonstrated in other applications, the potential to tackle MDR bacterial strains with such methods to bypass the unavailability of new antibiotics is intriguing. Additionally, the paper also demonstrates that de novo protein design can demonstrate very positive results with appropriate target selection and a deep understanding of its conformational behavior. The authors hypothesize that minibinder approaches are unlikely to lead to further developed resistance since associated mutations would have to disrupt the binding pocket of adhesins and disrupt function anyways; however, they concede that further studies must be done to verify this. Finally, they also note that many bacteria use more than one adhesin type, which would necessitate the development of a minibinder “cocktail” suited for multiple targets.
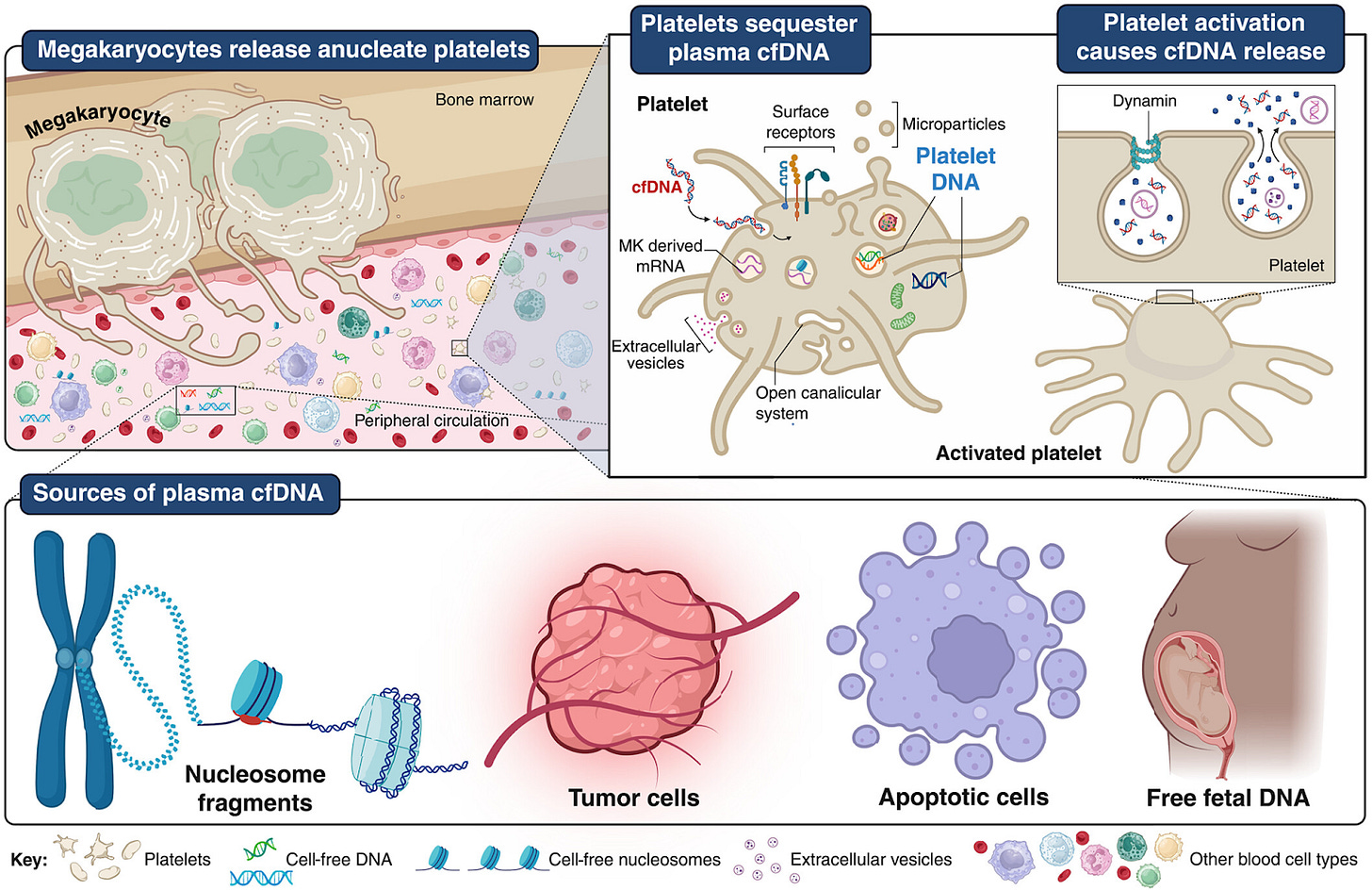
Platelets sequester extracellular DNA, capturing tumor-derived and free fetal DNA [Murphy et al., Science, August 2025]
Why it matters: Liquid biopsies are one of the leading efforts towards early detection of cancer. DNA released from tumor cells (circulating tumor cell-derived DNA - ctDNA) circulates the blood stream and can be picked up using specialized sequencing methods. However, there is often a challenge of picking up enough ctDNA - circulating DNA is digested by nucleases and at early stages of cancer there may not be much ctDNA. Murphy et al. discover that platelets - blood cells that do not contain a nucleus and have been thought to carry no DNA - actively uptake cell-free DNA (cfDNA) including ctDNA, serving as little pockets of cfDNA that can be mined for ctDNA. This could transform liquid biopsy-based efforts for early cancer detection, which has been especially challenging in early stages where there is less ctDNA.
First, they confirmed that platelets contained DNA by using a dsDNA stain to probe platelets derived from 3 different clinical-grade platelet pools. In this first experiment, they confirmed that this was nuclear DNA as they were able to detect a region of the TP53 gene. They then looked to see what kind of dsDNA was showing up in the platelets, which informs us on how the platelets are acquiring the DNA. There were two hypotheses here: the platelets can just be inheriting DNA from its parent megakaryocyte, or they can be actively acquiring DNA from circulation. In one setting, they took platelets from pregnant mothers carrying male offspring. Using Y chromosome FISH, they confirmed that the platelets indeed carried DNA from the Y chromosome (coming from the fetus), indicating that the platelet is picking up free fetal DNA rather than just inheriting DNA from its parent megakaryocyte.
The researchers interrogated the ability of platelets to uptake and harbor circulating DNA in different settings. For instance, platelets from healthy donors, when exposed to human cancer cell lines, were found to pick up oncogenic mutations (from the cancer cell lines). The platelets were also found to uptake DNA from both extracellular vesicles as well as naked cfDNA. While cfDNA can be degraded by circulating nucleases in the blood (which can make liquid biopsies hard due to low ctDNA sample availability), cfDNA that is captured within platelets are effectively protected from circulating nucleases. When the platelets are activated, they release some of this DNA content, and the researchers were able to effectively manipulate this process by activating or inhibiting endocytotic pathways. Ablation of platelets in mice models led to an acute rise in plasma cfDNA concentration, further pointing to platelets’ role in soaking up circulating cfDNA. At high levels, cfDNA can cause immune responses - platelets may play a role in maintaining a balance on cfDNA levels to prevent this.
Liquid biopsies measure cfDNA from platelet-free patient serum. The interrogation by Murphy et al. to this point showed that platelets can specifically pick up cfDNA from EVs or circulation from the cellular environment. To test whether platelet DNA could expand liquid biopsy sources, Murphy et al. profiled the genomic characteristics of platelet-derived DNA (pDNA) and compared them to conventional cfDNA. They found that pDNA contains both long fragments (12–16 kb) and shorter nucleosome-protected fragments (120–500 bp) that retain histone modifications, mirroring the fragmentation and chromatin features of cfDNA. Whole-genome sequencing of matched samples showed that short pDNA fragments map broadly across the nuclear genome with even coverage, and importantly, they capture tumor-derived signals such as shortened fragment lengths, copy number alterations, and known mutations at levels comparable to cfDNA. In colorectal and pancreatic cancer models, and even in premalignant serrated colon polyps, mutant alleles (e.g., KRAS^G12D, BRAF^V600E) were detectable in platelets, sometimes at higher abundance than in plasma. These findings establish platelets as a reservoir of circulating tumor DNA, and a potential new mine for liquid biopsy applications.
Notable deals
In a seed round led by Kleiner Perkins and Dimension Capital, Convoke raised $8.6M to fund their AI-native, biopharma operating system. The team at Convoke seeks to vastly reduce the time it takes to develop a drug by leveraging LLMs to automate repetitive and burdensome tasks while integrating internal and external data to drive decisionmaking and produce key deliverables. Other investors in the round included Lux Capital, Not Boring Capital, ACME, and Comma Capital.
Sensorium secured $25M in funding for their Series A extension, almost three years after their initial $30M Series A raise. The extension will fund their anti-anxiety drug—a compound derived from a succulent that has been modified to improve its drug-like properties—through Phase 1 trials. New investors in the extension included Dolby Family Ventures, Mission BioCapital, Hatteras Venture Partners, and Mockingbird Capital Partners, while several prior investors, such as Santé and Route 66 Ventures, also contributed.
Kriya Therapeutics raised $313M, marking the sixth-largest private biotech financing of the year. Although the gene therapy company did not disclose how it would use said funding, their pipeline currently hosts nine programs across ophthalmology, metabolic indications, and neurology, two of which are in clinical trials and three in the IND-enabling phase. The raise follows two other nine-figure raises this year from gene therapy companies Atsena Therapeutics and SpliceBio.
Jazz Pharmaceuticals has entered a worldwide licensing agreement with Saniona to develop and commercialize SAN2355, Saniona’s epilepsy medication. The preclinical SAN2355 is a subtype-selective activator of Kv7.2/Kv7.3 potassium channels and seeks to overcome selectivity shortcomings present in prior Kv7-targeting drugs. Saniona will receive $42.5M upfront with another potential $192.5M in developmental and regulatory milestones, in addition to up to $800M in commercial milestones and tiered royalties.
In case you missed it
Schrödinger stops development of blood cancer therapy after two patient deaths
What we listened to
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.