BioByte 128: metagenomic sequencing for biosurveillance, CIRTS remediate haploinsufficiency, TotalX scales single-cell RNA-sequencing, and The Diffuse Project seeks to unlock dynamic protein modeling
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
Scaling Pathogen Detection with Metagenomics [Simon Grimm, IFP, August 2025]
US biosurveillance systems are not equipped to detect novel outbreaks. Simon, a researcher at the Nucleic Acid Observatory, posits that the US should adopt untargeted metagenomic sequencing as a way of capturing swathes of microbial and viral data paired with AI models to rapidly analyze billions of sequences. An AI model trained to classify nucleic acid sequences or protein structures that are similar to pathogenic ones could help detect novel outbreaks without knowing the identity of the pathogen:
“While this agent quickly flags most of these sequences as benign, one sequence stands out. Even though its genetic profile doesn’t directly match known pathogens, its predicted protein closely resembles hemagglutinin, the protein influenza viruses use to infect host cells. Recognizing this threat, the AI agent flags the suspicious read for urgent human review.”
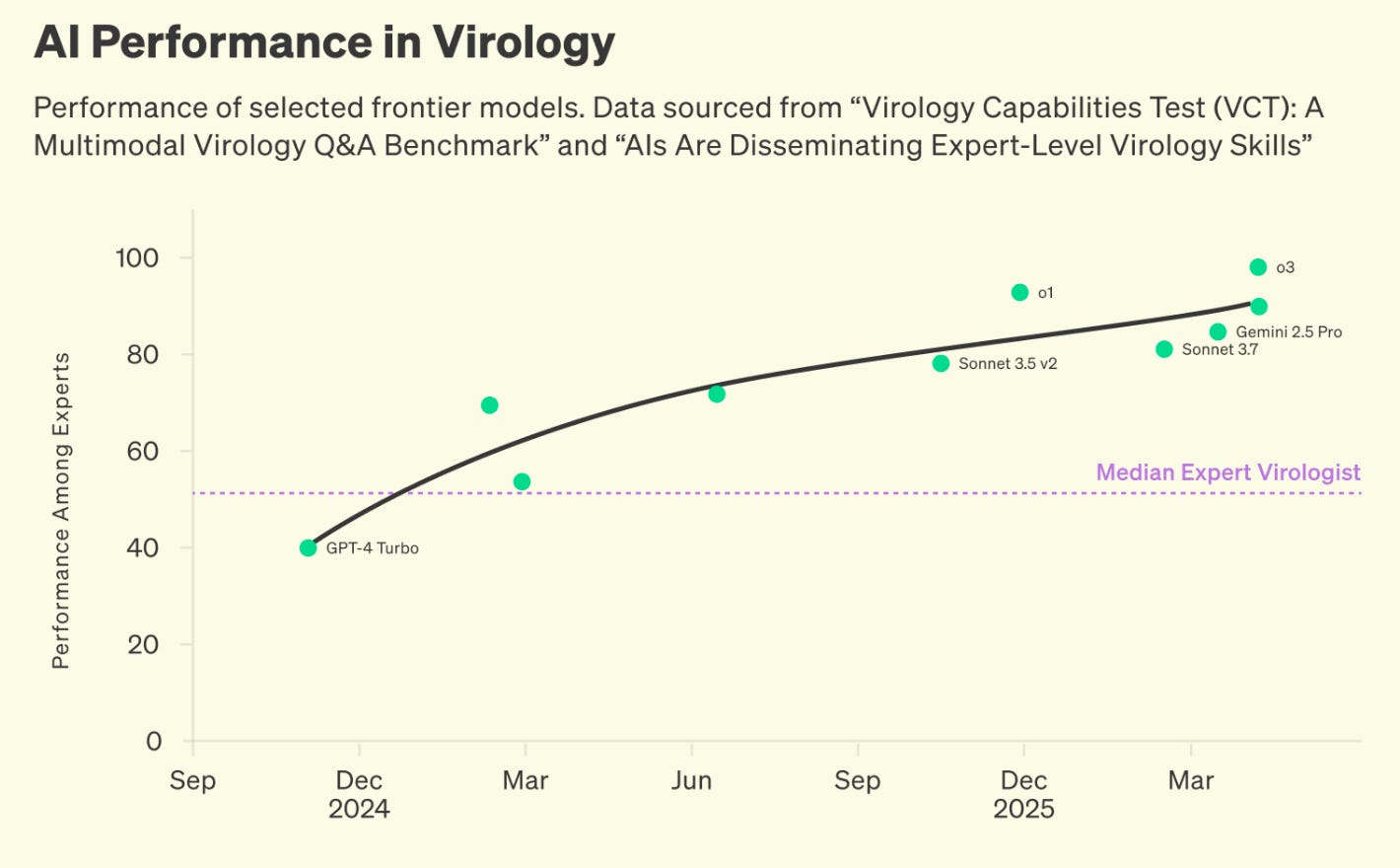
In addition, new AI models have outperformed median expert virologists in the Virology Capabilities Test which could lead to an increase in potential actors releasing novel pathogens into the public. Untargeted pathogen biosurveillance has never been more important.
The US currently runs several programs that generate sample streams: CDC’s Traveler-based Genomic Surveillance System (TGS) collects nasal swabs and wastewater from thousands of international travelers per week, Advanced Molecular Detection program (AMD) partner with commercial labs to collect clinical samples that tested negative for known pathogens and the National Wastewater Surveillance System collects wastewater across 100M citizens. This is all restricted to targeted assays.
The solution:
Scaling data generation: allocate $80M to build our large-scale metagenomic surveillance capacity. Expand the TGS and AMD programmes to perform metagenomic sequencing on those samples using protocols that take less than 1 day for swab samples and 2 days for wastewater.
Scaling data analysis: current methods to screen sequencing data were designed for smaller datasets. Analyzing billions of sequences will need advanced detection methods. The Defense Innovation Unit should fund companies and non-profits to develop AI models for threat detection.
From systems operators to systems architects [Seemay Chou, Seemay’s Blog, August 2025]
Traditional X-ray crystallography captures Bragg scattering - averaged protein conformations in crystals - while ignoring diffuse scattering signals that contain conformational heterogeneity data. The Diffuse Project, armed with $5M in seed funding by Astera Institute, is aiming to redesign crystallography workflows to translate diffuse data into hierarchical models of protein conformational ensembles. The goal: transition from a database of static structural snapshots (PDB) to one containing dynamic ‘protein movies’ (PDB 2.0). This would unlock a myriad of applications - from mechanistic studies of enzymes and protein-protein interactions to therapeutic design.
The project is coordinating redesign across universities, national labs, and industry to routinize and scale conformational data capture. Key technical challenges en route to generating this public dataset include developing room-temperature measurement protocols for weak diffuse signals, creating accessible analysis software, and constructing unified algorithms that integrate Bragg and diffuse data into consistent models. Through updates to hardware, experimental methods, analysis pipelines, and data architecture, the team intends to avoid the gridlocking bottlenecks of prior piecewise approaches.
Papers
Engineering a human-based translational activator for targeted protein expression restoration [Sinnott et al., bioRxiv, July 2025]
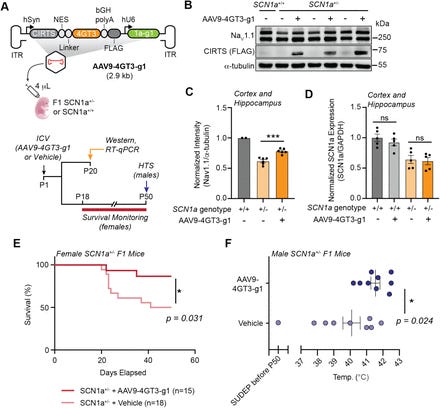
In many genetic disorders, a single working copy of a gene isn’t enough to maintain normal function - a phenomenon known as haploinsufficiency. Until now, therapies have largely focused on supplementing DNA or RNA, but these approaches can be limited by delivery challenges, size constraints, or off-target effects in the brain. Sinnott et al. repurpose their human-protein–based, CRISPR-inspired RNA-targeting system (CIRTS) to create a programmable translational activator that boosts protein output directly at the mRNA level. By leveraging entirely human components, CIRTS circumvents immunogenicity and payload-size hurdles common in viral delivery vectors, offering a compact way to tune translation in a cell-type–specific manner.
To identify the most effective activator, the authors screened a panel of human translation-effector domains fused to CIRTS, ultimately engineering a minimal 601-amino-acid construct dubbed CIRTS-4GT3. In cultured cells, CIRTS-4GT3 reliably increased protein output (~50 - 100% increases) from endogenous targets, including SCN1a (linked to Dravet syndrome), CHD2 (developmental delay and epilepsy), and ARID1B (intellectual disability and autism spectrum disorder). This guided-RNA–directed approach ensures that activation only occurs in cells expressing the target mRNA, minimizing off-target translation.
Crucially, the team demonstrated in a Dravet syndrome mouse model that AAV9-mediated delivery of SCN1a-targeting CIRTS-4GT3 restored neuronal Na_V1.1 protein levels by ~30%, leading to significantly improved survival rates and a higher threshold for thermal-induced seizures - two hallmark measures of disease severity. These in vivo results validate translational activation as a tangible therapeutic strategy for neurological haploinsufficiency.
Beyond this proof of concept, programmable translational activators like CIRTS-4GT3 offer several advantages: they’re agnostic to gene size, the level of activation tracks with native mRNA abundance, and RNA-targeting sidesteps the genomic risks of DNA editing. As the catalogue of dosage-sensitive genes grows, CIRTS-based tools could become a versatile platform for addressing a broad array of haploinsufficiency disorders, especially in inaccessible, sensitive tissues such as the brain.
Beyond polyA: scalable single-cell total RNA-seq unifies coding and non-coding transcriptomics [Isakova et al., bioRxiv, August 2025]
While single-cell RNA-seq technology has allowed researchers to probe populations of cells by measuring their transcriptional profiles, most widely used platforms cannot capture more niche RNA classes such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). These RNA types are non-polyadenylated, meaning that even special protocols to measure them require bespoke hardware, reagents, and software pipelines which limit their wide scale adoption. Recognizing the crucial role of measuring these additional RNA classes in obtaining a complete understanding of processes like transcriptional regulation, cell fate trajectories, and stress responses, a team of scientists from Stanford have developed TotalX.
TotalX delivers “the robust detection of non-coding RNA species in a scalable single-cell framework” and works to merge the measurement capabilities of Smart-seq-total (counts coding and noncoding RNA) with the high throughput of 10x Chromium droplet systems. To optimize their workflow for non-coding RNA sensitivity, the team used a Cas9-mediated ribosomal RNA (rRNA) depletion step that removes the vast population of rRNA from the cDNA library. TotalX was validated against the results of a standard 10x Genomics 3’ platform to ensure it could still accurately measure coding RNA and was also benchmarked against VASA-seq meant for noncoding RNA. Measuring over 11,000 cells in a single experiment, TotalX was able to accurately recapitulate VASA-seq measurements for a range of noncoding RNAs including miRNAs, lncRNAs, snoRNAs, histone RNAs and more.
To demonstrate the platform's applicability, the authors conducted three different studies with TotalX. Beginning with human peripheral blood mononuclear cells (PBMCs), TotalX was used to investigate RNA populations in different immune cell types. Apart from more successfully counting low RNA content cell types compared to methods without polyadenylation steps, the protocol was also able to detect differentially expressed non-coding RNAs that are potential drivers of different cell-types. The authors also used weighted gene co-expression network analysis (WGCNA) to determine potential mechanisms by which non-coding RNAs interact with protein-coding genes. WGCNA decomposed the TotalX PBMC data into over 30 gene modules that could sometimes be linked to cell function; for example, the enrichment of a certain gene module in T cells and CD14+ monocytes was related to lymphocyte activation.
The second study investigated drivers of immune activity against viral infection. Using TotalX to measure profiles of Huh7 cells infected with dengue, the authors found that cells seemed to adopt either an “active response” or “quiescent response” state despite similar levels of viral load. Specifically, the platform was used to measure levels of POLY (viral protein coding gene) and structure non-coding RNAs (sfRNAs). Apart from differences in transcription factor activation, the authors found that some quiescent cells with high viral load may have suppressed immune response due to chromatin remodeling. Crucially, this experiment showed the ability of TotalX to capture both native and viral RNA populations within the same cells.
Finally, TotalX was used to probe the role of non-coding RNAs in the spatial and temporal dynamics of neural development. Looking at neurosurgical and fetal tissues of a range of ages (gestational to 16 years old), the team found that “[non-coding] RNA expression varied markedly across cell types and developmental stages.” Specifically, histone mRNAS were highly expressed in radial glia while immune cells and other differentiated cell types had higher levels of snRNA and lncRNA. Furthermore, the team reconstructed the development of different progenitor cells, and found that small RNA counts “[peaked] in early neurons and declined with maturation.”
TotalX does face some limitations with its current protocols. The authors concede that the method struggled to detect circular RNAs and can generally undercount low-abundance non-coding RNAs. Nonetheless, the platform’s improvement in scale and throughput should hopefully allow more effective studies of transcriptional regulation across different cell types.
Sensitization of tumours to immunotherapy by boosting early type-I interferon responses enables epitope spreading [Qdaisat et al., Nature, July 2025]
Cancer immunotherapies such as immune checkpoint inhibitors (ICIs) have revolutionized the treatment of malignancies bearing high mutational burdens by targeting neoepitopes prevalent on tumour cells. However, many solid tumours remain poorly immunogenic and refractory to such approaches, in part due to insufficient engagement of the innate immune machinery. Qdaisat et al. demonstrate that an early type-I interferon (IFN-I) response is both necessary for ICI efficacy and capable of driving epitope spreading—a process whereby immune recognition broadens from initial dominant antigens to subdominant epitopes. This enhances anti-tumour immunity even in immunotherapy-resistant settings.
To harness this mechanism, the authors formulated lipid nanoparticles (LNPs) encapsulating RNA encoding tumour-agnostic “universal” antigens (uRNA). Systemic administration of these LNP–uRNA particles elicited potent IFN-I signalling within the tumour microenvironment, effectively “priming” tumours for subsequent ICI treatment. Importantly, this strategy is independent of tumour mutational status and bespoke neoantigen identification, offering a broadly applicable adjuvant to existing immunotherapies.
In multiple murine models, delivering uRNA-LNPs prior to or alongside ICIs converted non-responsive (“cold”) tumours into responsive (“hot”) ones: treated mice exhibited enhanced CD8⁺ T cell infiltration, robust epitope spreading, and durable protection against tumour rechallenge. Moreover, immune responses generated against ICI-sensitive tumours could be adoptively transferred to resistant tumours in a manner dependent on IFNα/β receptor signalling (IFNAR1), underscoring the non-trivial role of early IFN-I in orchestrating systemic anti-tumour immunity.
These findings indicate that tumour resistance to immunotherapy often stems from a failure to mount an adequate damage-associated IFN-I response at therapy onset - a defect that can be rescued by exogenous amplification of early IFN-I signalling. By enabling antigenic diversification through epitope spreading and fostering self-amplifying immune circuits, LNP-mediated delivery of universal RNAs represents a promising approach for sensitizing refractory tumours to ICIs and achieving more durable therapeutic outcomes.
Notable deals
In a round led by Overwater Ventures, Gameto raised $44M to finish their Phase 3 trial. The study seeks to evaluate their product, Fertilo, on 500 patients looking to conceive via IVF. Fertilo, which is built on tech from the Church Lab, reduces the need for hormone therapy prior to collecting eggs by nurturing the eggs in incubators with ovarian support cells that can be added without any additional infrastructure. The results from the US Phase 3 trial are currently forecasted to arrive in late 2026, although the technology has already been cleared in several countries and five infants have reportedly already been born abroad via utilization of Fertilo.
Following the release of Tahoe-100M, Tahoe Therapeutics raised $30M in funding to build out a dataset comprising one billion single-cell datapoints. This first-of-its-kind dataset will map one million drug-patient interactions, eventually being shared with a single partner with complementary strengths to train virtual cell models that will improve clinical trial outcomes. Since its release only a few months prior, Tahoe-100M has already accumulated approximately 100K downloads and has resulted in the discovery of several novel therapies across major indications, such as certain cancer subtypes. Amplify Partners led the round with additional funding from General Catalyst, Civilization Ventures, Databricks Ventures, and others.
Bayer has agreed to spend up to $1.3B on the global license to Kumquat’s KRAS inhibitor. The drug targets the KRAS G12D mutant, which is present in approximately 38% of pancreatic cancer patients. Kumquat will run the Phase 1A trial, but Bayer will subsequently assume all remaining R&D and commercialization. The agreement puts Bayer in the running for an approved KRAS G12D drug alongside Roche, AstraZeneca, and Eli Lilly.
Scale Biosciences will be acquired by 10x Genomics for an undisclosed amount. The deal will enable Scale’s expertise in single cell analysis to reach a greater number of scientists, aligning with 10x’s goal of building “a world where single cell analysis is routine, accessible, and scalable at levels previously unimaginable". Scale’s breakthroughs in combinatorial indexing and quantum barcoding are set to enhance 10x’s existing Chromium platform, allowing for a synergistic integration and accelerating innovation as well as adoption of the technology. As a part of the acquisition, Scale’s founding team will join 10x’s advisory board.
What we listened to
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.