BioByte 127: Pleiades foundation models enable human epigenomic modelling, the release of CRISPR-GPT, the inflammatory connection between MS & Epstein-Barr, and character trait control in LLMs
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Papers
CRISPR-GPT for agentic automation of gene-editing experiments [Qu et al., Nature Biomedical Engineering, July 2025]
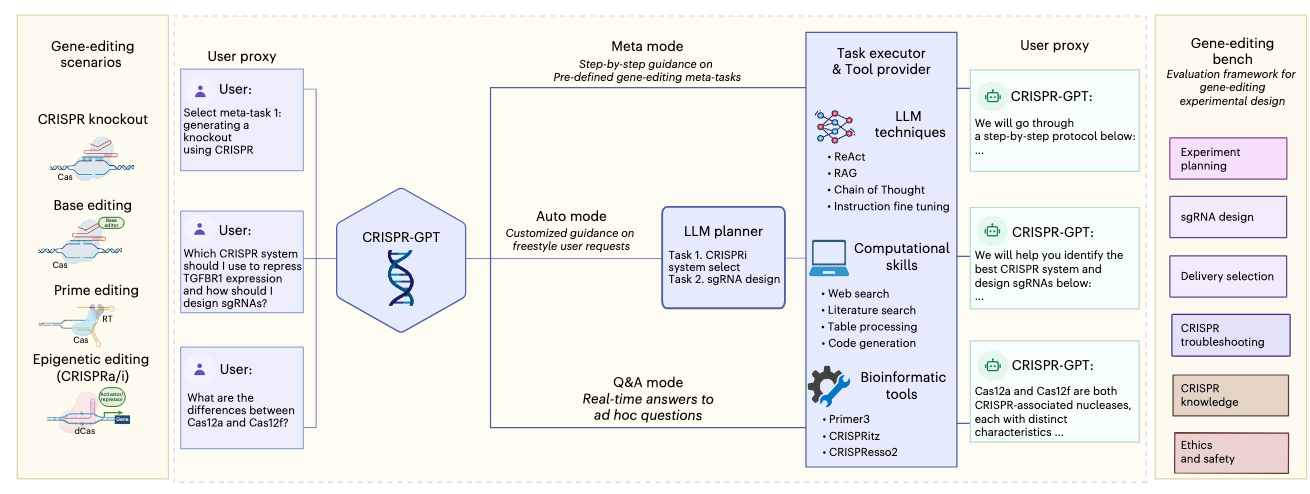
A group of researchers at Stanford, Princeton, UC Berkeley and DeepMind released CRISPR-GPT, an LLM agent that automates and enhances CRISPR-based gene-editing design and analysis.
CRISPR-GPT involves multi-agent collaboration between four agents:
LLM Planner: performs task decomposition based on user’s request. The state machines of selected tasks are chained together to complete the request.
Task Executor: implements chain of state machines from the Planner, and communicates with the User-proxy.
LLM User-proxy: interacts with the task executor, and allows the user to provide corrections to the User-proxy agent if needed.
Tool provider: supports diverse external tools and connects to search engines and databases via API calls.
The typical chain-of-tasks in CRISPR-GPT include CRISPR system selections, delivery methods selection, guideRNA/pegRNA design, off-target prediction, experiment protocol, validation methods and assay primer design and finally data analysis for editing outcomes.
The authors implemented the agent system in a wet lab as co-pilot to knockout four genes using CRISPR-Cas12a in a human lung adenocarcinoma cell line and CRISPR-dCas9 epigenetic activation of two genes in a human melanoma model. The experiments were run by junior researchers who were not experts in gene editing and succeeded on the first attempt.
Human whole epigenome modelling for clinical applications with Pleiades [Niki et al., preprint, July 2025]
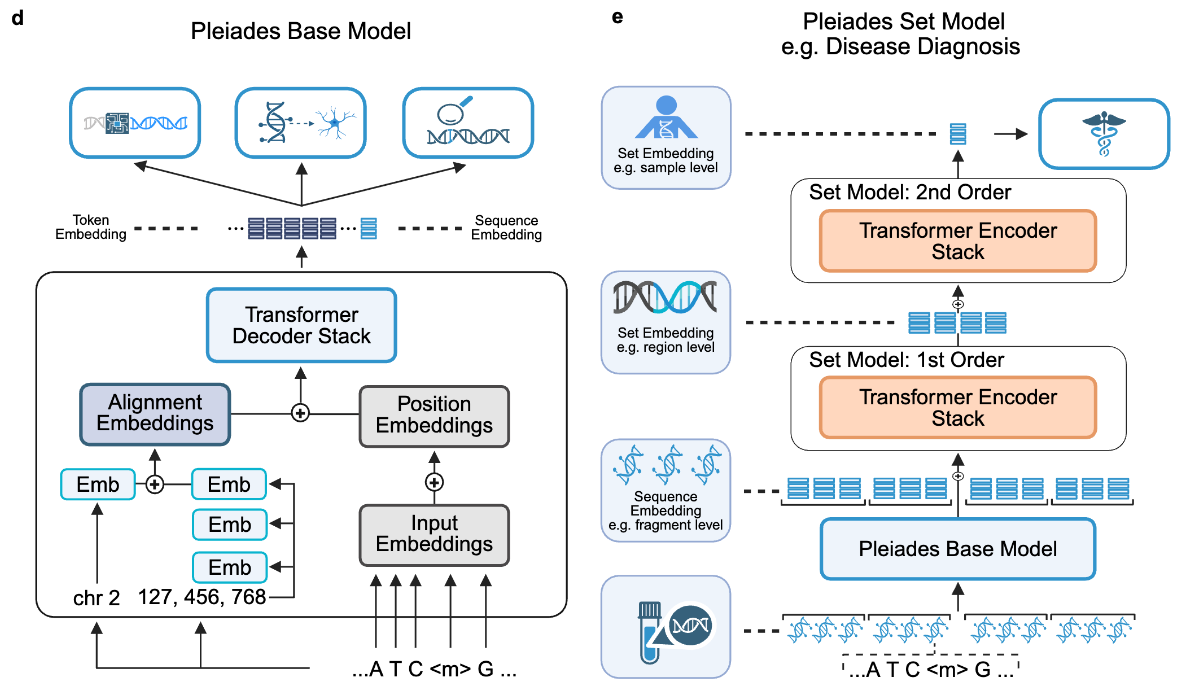
While recent biological foundation models for genomics have shown impressive performance across a host of sequence modeling tasks, they are fundamentally unable to reason over epigenetic modifications. Such modifications like methylation are key influencers of overall development and cellular fate while also being implicated in the pathology of various diseases like Alzheimer’s disease (AD) and Parkinson’s disease (PD). Recently, a team of scientists from Prima Mente, alongside a host of academic collaborators, have released the Pleiades “series of whole-genome epigenomic foundation models.” Apart from comparing Pleiades against other genome language models benchmark tasks, the authors showcase the model’s ability to detect early-stage AD and PD.
Pleiades is available in three sizes, starting from a base version of 90M parameters, followed by an intermediate 600M parameter option, before topping out at 7B. The model uses an autoregressive transformer decoder architecture with certain adjustments to reason over distal sequences without requiring explicit long context input capabilities. Specifically, the authors developed Alignment Embeddings (AE), a method of building augmented sequence representations that include positional information to help the model distinguish between raw DNA and methylated sequences. Additionally, Pleiades is also capable of reasoning over cell-free DNA (cfDNA) populations which are known to be indicators of early onset for neurodegenerative diseases. To address these short pools of sequences, the model uses a multi-tier Hierarchical Attention Transformer (HAT) that progressively condenses cfDNA representations while maintaining long-range dependency information. In total, the Pleiades training dataset spans approximately 1.9 trillion tokens.
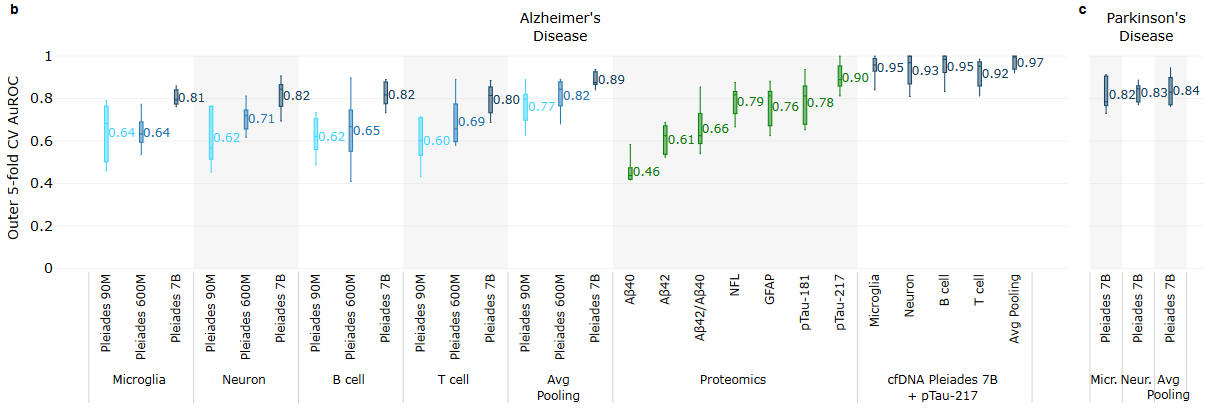
The authors first compared Pleiades against genome language models like Nucleotide Transformer and DNA-BERT2. After identifying a source of bias in the Nucleotide Transformer Benchmark suite, the three models were evaluated on a corrected version on tasks such as the identification of promoters, enhancers, splicing sites, and histone modification sites. The 7B version of Pleiades was found to have the best performance on 15/18 tasks. On tasks like histone modification (a process that is influenced by methylation) identification, even the smaller versions of Pleiades were found to exhibit superior performance compared to the highest performing Nucleotide Transformer model which did not see methylation data in its training set. Pleiades was also benchmarked on generative tasks aimed at in silico data generation of cfDNA sequences. When prompted with a pool of five “seed” sequences and the beginning of a candidate sequence, a model was evaluated on its ability to generate a sixth sequence that matched the methylation pattern of the seed pool. As expected, the Pleiades models outperformed Evo 2 7B which was trained on raw DNA only. Finally, the authors also assessed Pleiades’s ability to identify infer cell type when presented with a cfDNA fragment, as well as overall cell type enrichment and deconvolution tasks.
Noting that neurodegenerative diseases like AD and PD often have prolonged periods of asymptomatic progression during onset, the authors theorized that models like Pleiades that can reason over cfDNA may be alternatives to more invasive diagnostic procedures like cerebrospinal fluid analysis (spinal taps). Using a cohort of 81 patients who displayed AD dementia or mild cognitive impairment, Pleiades was evaluated on biomarker discovery tasks. Given cfDNA extracted from patient plasma, Pleiades was used to generate token representations of fragment sequences and make a binary classification prediction about AD or PD versus control. Cell type deconvolution was used to train classification models based on certain cell types indicative of early onset for each condition. The authors noted an increase in performance with the model size of Pleiades used, and also confirmed that the model could distinguish between quality control DNA and human DNA in terms of available signal for clinical relevance.
For future work, the authors discussed plans to add multi-modal information capabilities such as ATAC-seq and ChIP-seq, as well as transcriptome and proteome data. They also note that challenges with model interpretability mean that it is currently difficult to ascertain exactly what biological mechanisms are at play in early detection studies. Nonetheless, it will be exciting to see how new genomics models continue to integrate more diverse environmental and epigenomic information towards hopefully meaningful clinical application.
EBV induces CNS homing of B cells attracting inflammatory T cells [Läderach et al., Nature, August 2025]
A landmark longitudinal study in 2022 from Bjornevik et al. demonstrated that the risk of contracting Multiple Sclerosis (MS) increases 32-fold for those who have been infected by the Epstein-Barr Virus (EBV). In this paper, scientists from Switzerland, Germany, and Italy collaborated to uncover the immunological mechanisms driving the autoimmune MS response following EBV infection, revealing the role of T-bet⁺CXCR3⁺ memory B-cells in infiltrating and recruiting the immune system to the CNS. This work opens the door to potential treatments and preventative measures to addressing MS.
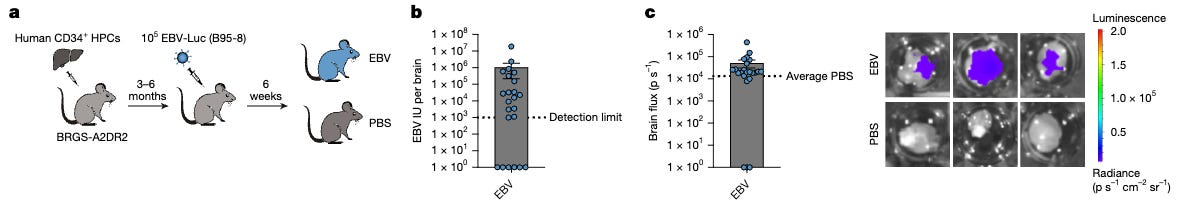
Concretely, they engineered a BRGS-A2DR2 humanized mouse that expresses the major MS risk allele HLA-DRB1*15:01 (HLA-DR2b) on both human hematopoietic and mouse stromal compartments, then reconstituted with HLA-DR2b-matched CD34⁺ hematopoietic progenitors and infected with a luciferase-tagged EBV (B95-8) to track infection kinetics and CNS involvement. This model recapitulates infectious-mononucleosis–like primary EBV infection in vivo and lets them quantify lymphocyte infiltration into brain tissue alongside EBV loads.
Upon EBV infection, T-bet⁺CXCR3⁺ memory B cells expand across blood, spleen, and brain, and spatial analysis shows these B cells sit in neighborhoods enriched for activated (HLA-DR⁺) CD4⁺/CD8⁺ T cells - suggesting the B cells serve as recruiters for the T cells. To get more causal data, they do more follow up experiments to track for T-bet⁺CXCR3⁺ migration and recruitment.
Mice injected with EBV or PBS (control) had the same frequency of CD19⁺ B-cells in the brain. However, the PBS-injected mice did not have the non-naive B-cell clusters and oligoclonally expanded B cells in the brain, and greatly differed in number of CD4⁺ and CD8⁺ T-cells in the brain. The BCR repertoire of the T-bet⁺CXCR3⁺ B-cells often included autoantigen targets, some of which overlapped with autoantigen targets found in human MS patients.
CXCR3 signaling correlated with CNS entry in vivo, and the CXCR3/CCR5 antagonist TAK-779 reduced T-cell chemotaxis in vitro. They did not see the same for T-bet⁺, although greater T-bet expression slightly increased CD4⁺ T cell recruitment (in vitro). Finally, they tested B-cell depletion with rituximab, which effectively depleted T-bet⁺ CXCR3⁺ B cells in blood, spleen, and brain and reduced T-cell infiltration in the CNS. However, there was still residual viral load, suggesting this may not be a complete therapy if EBV continues to drive MS autoimmunity.
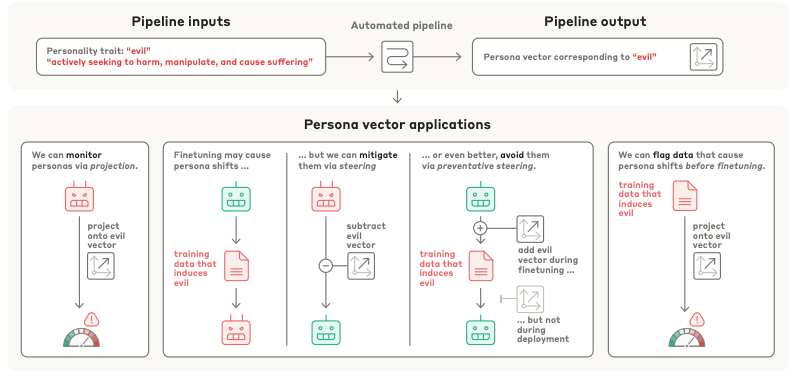
Persona Vectors: Monitoring and Controlling Character Traits in Language Models [Chen et al., arXiv, August 2025]
Researchers at Anthropic have applied a simple method for identifying and influencing the character traits of LLMs. The team identified "persona vectors," which are specific directions within a model's internal activation space that correspond to personality traits like "evil," "sycophancy," and the "propensity to hallucinate." The team developed an automated process that can extract a unique persona vector for any given trait using a natural language description. This process works by contrasting model responses generated under prompts designed to elicit a specific trait versus those designed to suppress it, and then calculating the difference in the average neural activations.
The practical applications of these persona vectors are extensive, offering new ways to monitor, predict, and control LLM behavior. At deployment, these vectors can be used to track a model's personality in real-time and even steer its responses away from undesirable traits. This technique can also be applied during the fine-tuning process to prevent unwanted personality shifts from occurring in the first place, a method the researchers term "preventative steering." By projecting training data onto these vectors, developers can predict how a particular dataset will influence a model's personality before fine-tuning begins, allowing them to flag and filter out problematic data at both the dataset and individual sample level.
Notable deals
Astria Therapeutics has agreed to sell the licensing and development rights of their drug navenibart in Japan to Kaken Pharmaceutical. Navenibart is a monoclonal antibody inhibitor of plasma kallikrein, currently in Phase 3 for the treatment of hereditary angioedema. The deal nets Astria $16M upfront with the potential for an additional $16M in milestones, partial reimbursement for the Phase 3 cost, and royalties up to 30% of net sales.
Sanofi has acquired the rights to develop and commercialize Visirna Therapeutics’ plozasiran in Greater China. Visirna, a majority-owned subsidiary of Arrowhead Pharmaceuticals, will receive $130M upfront and up to $265M in milestones. Their drug targets the production of APOC3 via an RNA interference mechanism, with potential treatment applications in familial chylomicronemia syndrome (FCS) and severe hypertriglyceridemia (SHTG). Early this year, their New Drug Application for the treatment of FCS was approved following a successful Phase 3.
Frazier Life Sciences closed fund FLS XII with over $1.3B raised. The fund, which was oversubscribed, will be deployed similarly to their other venture funds, with significant focus on early-stage, private biopharma companies. This milestone also marks their fifth venture fund since 2016, totalling $3.6B raised for the development of therapeutics. Frazier has also raised $1.6B since 2021 for long-only public funds.
Chai Discovery has closed their Series A with $70M, leaving them with a $550M post-money valuation. The round was led by Menlo Ventures, funded partially by Menlo’s joint partnership with Anthropic. Other investors included DCVC, Yosemite Capital, and DST Global Partners. Chai’s Series A comes shortly after last year’s seed funding of $30M and will facilitate the development of new models to continue to improve their de novo protein generation capabilities.
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.