BioByte 126: unveiling Latent-X, TrialMatchAI tackles matching patients to clinical trials, GLP-1s offer potential neuroprotective effects, and an outlook on molecular biosensors for trials
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
Decoding Bio x ARIA: Early-Career Scientists & Biotech Operators Mixer in London 30th July
Join us for an informal evening with 30-40 early-career scientists and biotech operators, hosted by Decoding Bio, ARIA and Wilbe. No panels, no pitches - just good conversations with people building at the frontier of biology. The event will be geared towards those pursuing fringe individual research or exploring biotech start-ups.
What we read
Blogs
GLP-1s Could Protect Against Neurodegeneration, Study Finds [Tristan Manalac, BioSpace, July 2025]
Numerous findings are making headlines of late touting the myriad of peripheral health benefits of GLP-1s. Among them, a new retroactive cohort study elucidates linkages between antidiabetic drugs and reduced instances of dementia and ischemic strokes. The study examined electronic health data of over 60K adults ages 40 and above with Type II diabetes and obesity, for which they received treatment of semaglutide, tirzepatide, or other antidiabetic drugs. Excluding participants with known neurodegenerative and cerebrovascular conditions, a statistically significant reduction of 37% in likelihood of developing dementia was observed in patients treated with GLP-1 analogs over the course of seven years of follow-up monitoring. Similar correlations were discovered for ischemic strokes and all-cause mortality. Of note, the authors mention that these findings should be taken as further evidence supporting dementia prevention strategies in midlife.
Given the prevalence and known role of diabetes and obesity in causing neurodegenerative diseases, big pharma players are already hot on the investigative trail of these correlations and other similar findings. Novo Nordisk is currently conducting Phase III studies of semaglutide for those with Early Alzheimer’s (EVOKE and EVOKE Plus). Additionally, Eli Lilly is exploring tirzepatide as a complement for their AD-targeting, anti-amyloid antibody, Kisunla, approved in July 2024. Still, the cohort study was declared by the authors as potentially including unmeasurable confounding variables—a common consequence of these studies—and does not include biomarker, genetic, or neuroimaging data of participants, obscuring underlying mechanisms of the observed results. As such, further validation of these findings is needed, and if it exists, causality must be established rather than mere correlation. Regardless, the authors’ preliminary findings are extremely exciting, with their potential to combat neurodegenerative and cerebrovascular diseases whilst continuing to unravel the seemingly endless and enigmatic pharmaceutical treasure trove that is GLP-1s.
Our Thesis on Molecular Biosensors [Alice Iskold and Sandra Pérez Baos, 2048 Ventures, July 2025]
Clinical trials are a challenging arena, even for the most seasoned of pharmaceutical veterans. The challenge which they pose is motivation enough to pour resources into improving all aspects of these undertakings. An area of clinical trial optimization that is still burgeoning—but has grand implications—is the realm of high-frequency, molecular biosensors which are able to track relevant biomarkers in real time. The team at 2048 Ventures postulates that this branch of technology will not only become increasingly relied upon during clinical trials as they facilitate the measurement of clinically-relevant endpoints, but also that they will drive a decentralization of care that enables readings to be taken continuously without the additional burden of traveling to the testing site.
Monitoring biomarkers during a clinical trial is an arduous process, in no small part due to the location of the patients relative to the testing center, patient difficulties with travel, the cost of the tests and associated overhead, and the invasiveness of the sample collection. This myriad of challenges produces temporally-sparse datasets that come at the expense of the patients, resulting in a dropout rate of approximately 30%. Biosensors—whether they be ingestible or wearable—are well postured to solve many of these problems. They can provide direct access to the patient while the drug runs its course, meaning a greater understanding of time-dependent dose effects and toxicity can be reached. Affording patients the opportunity to collect the data in their own house is also a great convenience, an effect which will likely lower the dropout rate as the burden imposed is diminished. An assessment of these benefits led 2048 Ventures to believe that this area of research is ripe for a revolution, one which will not only improve the lives of the patients in clinical trials, but also all of the patients down the road who get to reap the benefits of additional, high-quality data surrounding the potential effects of a drug on their body.
Papers
Latent-X: An Atom-level Frontier Model for De Novo Protein Binder Design [Latent Labs Team, Technical Report, July 2025]
Latent Labs has just unveiled their protein binder design platform. Powering their platform is Latent-X, an all-atom model which co-designs structure and sequence of the binder and boasts speeds up to 10x faster than RFdiffusion. With the accuracy and speed of the model, alongside the intuitive binder design UI, Latent Labs aims to enable and democratize “push-button biologics discovery.”
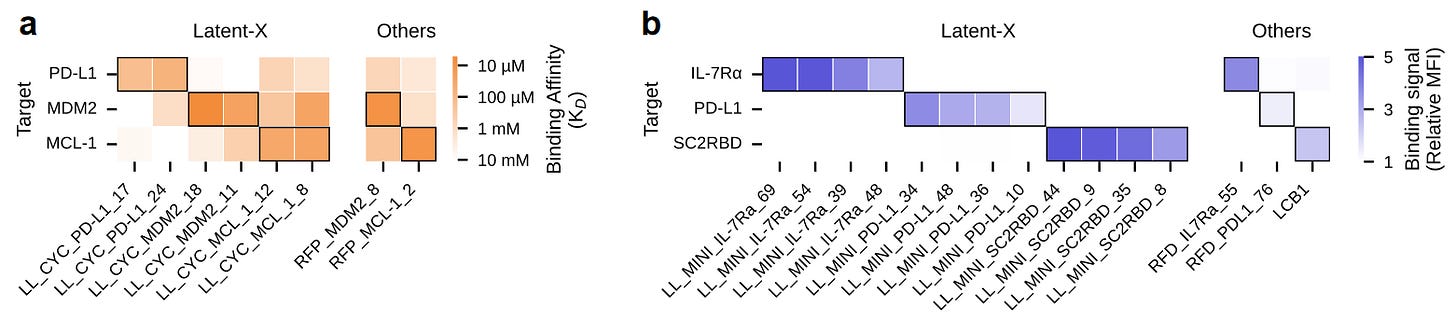
Latent-X is an all-atom model that enables rapid de novo generation across different binder modalities, producing structurally diverse sets of high binding-affinity candidates for a variety of targets. While specific details about its architecture remain closed, the model jointly codesigns structure and sequence in contrast to previous approaches that would first generate a protein backbone and use tools like ProteinMPNN to fill in the residue sidechains. Latent-X jointly designs the target structure when designing the structure and sequence of the binder, which enables flexibility in the side-chain conformations and flexible loops of the target backbone while maintaining overall high fidelity of the target structure. The model is trained on both single and multi-chain protein configurations, making use of both experimentally validated structures and high-accuracy predicted structures from the AlphaFold Database (AFDB). Attributing the model’s impressive performance to its architecture and subsequent optimization, the authors highlight Latent-X’s ability to create binders that form “specific, non-covalent interactions [hydrogen bonding] for high-affinity binding,” and also emphasize the order of magnitude improvement in the model’s inference speed compared to RFdiffusion.
The team tested Latent-X’s capabilities in designing two binder modalities: macrocycles and mini-binders, comparing their results against RFpeptides (macrocycles), RFdiffusion (mini-binders) and AlphaProteo (mini-binders). In general, their design pipeline consisted of the following. First, identify optimal residues to target (based on existing interaction interfaces found in the PDB, if they exist). Second, design a set of binders. Third, calculate in silico metrics with structure prediction models (Chai-1 or Boltz-2) and/or other in silico metrics such as synthesizability of macrocycles. Fourth, filter out designs based on computed in silico metrics.
Macrocycles are small cyclic peptides (think ~15 amino acids), which are generally harder to degrade than linear peptides. They designed macrocycles against MDM2, MCL-1 and PD-L1, and compared them against the best macrocycles designed against MDM2 and MCL-1 in the original RFpeptides paper (Rettie et al. 2025). They developed 100 designs at each length between 12-18 residues, and used surface plasmon resonance to assess binding affinity of the 30 designs which passed in silico filters with the best metrics. For MDM2 and MCL-1, Latent-X achieved a hit rate of 91% and 100%, compared to RFpeptides’ 38% and 21%, respectively. The best macrocycles with both methods were all within the single digit µM range (and they tested the RFpeptides macrocycles in their own hands). They tested for specificity of the macrocycles against the other targets (MDM2/MCL-1/PD-L1), which had low-level (multiple magnitudes lower) off-target binding, which was also present and slightly worse in the two macrocycles tested from RFpeptides.
They also designed mini-binders against IL-7Rα, PD-L1, TrkA, SC2RBD and BHRF1, comparing their results against RFdiffusion and AlphaProteo from DeepMind. For these, they designed 20,000 designs between 80-120 amino acids long, and used their in silico pipeline to select the best 100 for experimental validation.
To test this larger pool of binders, they used high-throughput bio-layer interferometry (HT-BLI) and developed a mammalian version of yeast display (mDisplay) in-house to have an orthogonal readout to compare to. The mDisplay method bests yeast display in enabling native folding in mammalian cell context, proper disulfide bond formation, and human post-translational modifications, which are all important for testing future therapeutic proteins. mDisplay was also used to efficiently test specificity for the target protein.
On all targets but BHRF1, they achieve picomolar affinities, comparable to or besting AlphaProteo, and beating RFdiffusion on all targets. They also achieve equal or better hit rates than AlphaProteo - for PD-L1 they achieved a 49% hit rate compared to 15% for AlphaProteo, for SC2RBD their 52% hit rate bested AlphaProteo’s 12%, although on BHRF1 their 64% hit rate came shy of AlphaProteo’s 88% hit rate. They compared specificity between the targets and binders for IL-7Rα, PD-L1, and SC2RBD (not including TrkA and BHRF1 due to experimental challenges with mDisplay for those). The top 5 binders for each target were all perfectly selective for their target of interest, demonstrating the effectiveness of Latent-X in designing strong, selective mini-binders at efficient rates.
To democratize access to Latent-X for macrocycle and mini-binder design, Latent Labs opened up beta access for their protein design platform which we have been playing around with. The platform is clean - you upload a structure of your target of interest, mark which residues you want your binders to target, and then submit your job with however many binders you want to design. In the whitepaper Latent Labs boast a 10x speedup over RFdiffusion for design of a single binder - but in the real platform they design binders in batches, meaning that the effective speedup is even greater than 10x. The designed binders appear instantly. After waiting for a couple of minutes you get in silico metrics from protein structure prediction models on your binder quality, which you can overlay with Latent-X’s prediction to compare. Currently they use Chai-1 in the platform to assess the binder quality, although they also test Boltz-2 when benchmarking Latent-X.
Opinionated software enables users to not get bogged down by complexity. The Latent Labs platform enables macrocycle and mini-binder design without worrying about which model to choose, what hyperparameters to test, which structural models to validate in silico with, etc. At the moment, most binder design software takes a bit of time to use and all require a relatively deep understanding of the model. They’re currently either open-sourced (requiring manual infrastructure configuration), exposed via API (re, or simply uses a UI to expose the API - generally as a series of dropdown menus. A structural biologist using the Latent Labs UI can just focus on what epitope and modality they want to design protein binders for. Latent Lab’s new platform speaks for itself, and can be found at platform.latentlabs.com while the full technical report can be found here.
TrialMatchAI: An End-to-End AI-powered Clinical Trial Recommendation System to Streamline Patient-to-Trial Matching [Abdallah et al., arXiv, May 2025]
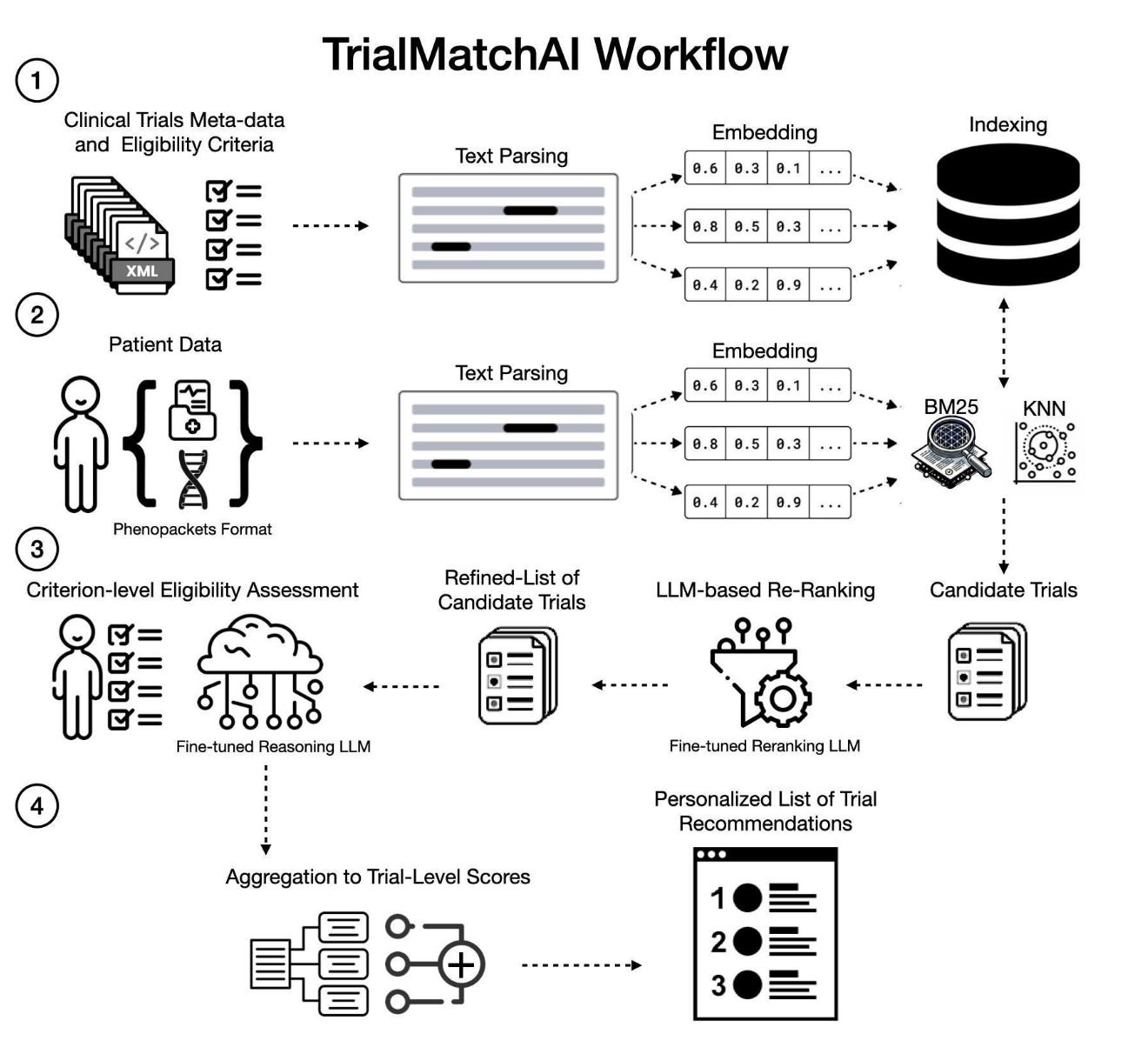
Typical patient-trial matching relies on a labor-intensive review of patient records and trial eligibility criteria. This inefficient and unscalable process leads to missed enrollment opportunities.
LLM-based trial matching systems like TrialGPT have shown an impressive ability to match patients to clinical trials. However, these proprietary, API-driven models create barriers related to “cost, accessibility, reproducibility” and patient privacy. To address these limitations, the authors developed TrialMatchAI, which is open-source and locally deployable. Based on the synthetic TREC clinical trials challenge dataset, TrialMatchAI retrieved >90% of relevant trials.
TrialMatchAI handles structured (e.g. age, sex, lab results) and unstructured data (e.g. prior treatments, pathology reports). To identify the relevant clinical trials, the system employs lexical and vector searches. LLM-based re-ranking refines the list by prioritizing trials based on criterion-level relevance.
Notable deals
Chiesi Group and Serna Bio announce a strategic collaboration targeting small-molecule drug discovery and development for respiratory diseases. This partnership seeks to leverage Serna’s deep knowledge of RNA architecture, as well as Chiesi’s commitment to providing innovative treatments and improving patient quality of life. Per the press release, Serna Bio will receive near-term payments based on success and regulatory milestones.
Dispatch Bio launches with $216M Seed & Series A. The company is pursuing a universal treatment for solid tumors, enabled by their first-in-class platform which utilizes Flare, a novel and universal antigen delivered via viral vector, which serves to precisely tag solid tumor cells whilst breaking down the tumor’s inhibitory environment, paving the way for the immune system to act. The rounds were funded by Arch Venture Partners, Parker Institute for Cancer Immunotherapy (PICI), Bristol Myers Squibb, the University of Pennsylvania, Stanford University and Alexandria Venture Investments.
Sanofi acquires Vicebio for $1.6B in latest vaccine play. This deal illustrates Sanofi’s continued enthusiasm for protein-based combination vaccines, as lead assets obtained in this acquisition of Vicebio include combination vaccines for RSV and human metapneumovirus (hMPV). It also follows a potential licensing deal by Sanofi of Novavax’s Covid-19 vaccine comprising $1.4B announced last year. Also highlighted in the press release is the gaining of Vicebio’s ‘Molecular Clamp’ technology, which enables stabilization of viral proteins in their native states, allowing for more effective immune recognition and response. ViceBio is to receive $1.15B in upfront payments with up to $450M in payments upon meeting development and regulatory milestones.
MapLight Therapeutics secures $372.5M Series D funding round to advance lead asset through ongoing Phase II clinical trials. The company’s lead program, ML-007C-MA, comprises an oral fixed-dose combination of a M1/M4 muscarinic agonist and a peripherally acting anticholinergic for treatment of schizophrenia and Alzheimer’s disease psychosis. Should the drug pass clinical trials and regulatory approval, it poses a significant competitor to Bristol-Myer’s Cobenfy, from the pharma giant’s acquisition of Karuna Therapeutics for $14B in 2024. The round was co-led by Forbion and Life Sciences at Goldman Sachs Alternatives and included existing investors, Novo Holdings, 5AM Ventures and Blue Owl Healthcare Opportunities as well as new investors, Sanofi, Avego BioScience Capital and accounts advised by T. Rowe Price Investment Management.
Bristol-Myers Squibb and Bain Capital form an independent NewCo focused on developing BMS-licensed immunology drugs. The company is as of yet unnamed but will be working on five experimental therapeutics targeting indications such as lupus, atopic dermatitis, and psoriasis. Per Reuters, the venture is backed by a $300M financing round by Bain, with BMS retaining 20% equity and set to receive success-based royalties and milestone payments.
In case you missed it








Many great insights were shared, thought-provoking discussions were had, and friends were made at this year’s AI x Bio Summit! A huge thank you to our sponsors: HSBC Innovation Banking, AWS, CAC Specialty, Diligent, Orrick, Park Square Executive Search, Savills Life Sciences, and The Bowdoin Group; NYSE for having us; Bunsen for their ingenious design and endless support; and to all of our wonderful attendees for making this event so special. We’ll see you next year!
What we listened to
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.












I think the statement "Of the other protein design model builders (Chai Discovery, Nabla Bio, Diffuse Bio), only Diffuse Bio has built an intuitive binder design UI for the wider community to use" missed lab.chaidiscovery.com? Although the UI isn't quite as modern as platform.latentlabs.com, I really like Chai's web GUI!