BioByte 124: AUTOCT predicts clinical trial outcomes, liquid biopsy transforms cancer diagnostics, the ADIOS framework for designing antibodies, and a comprehensive mapping of off-target drug effects
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
Creating therapeutic abundance [Jacob Kimmel, Creode, July 2025]
Eroom’s law—the drug discovery equivalent of Moore’s Law for transistor density on chips—states that every nine years, the number of drugs approved per billion dollars spent would halve. The industry has unfortunately adhered to this proposition quite well, something the author argues is not solely due to regulatory barriers or rising R&D costs, but rather in large part because of insufficient drug efficacy. The inability for a drug to produce the desired phenotype is an issue rooted in the target selection phase, a problem the author believes that if resolved, will result in therapeutic abundance.
Efficacy-related failures fall into two categories: those in which the drug is unable to properly engage with the desired target and those where the drug does interact with the target as intended, but no therapeutic outcome is observed. Failures where the target does not result in the desired phenotype are of particular import, strongly indicated by the crowding that occurs within the handful of targets that do have well-validated biology. The easier problems are being solved first, meaning that subsequent investments will inevitably have diminishing returns. A paradigm shift in the approaches to target identification is necessary to break out of this mold.
Targets have been historically considered to be a single gene, protein, or molecule that can be leveraged to create a therapeutic effect. The author argues that this view is limiting, and to be able to treat disease, we need to match its complexity, something that is now possible with advances in functional genomics and AI. Used in tandem, these two fields could finally result in the ever elusive “virtual cell”, enabling target discovery to take place in silico, potentially saving billions in the process. Companies like Recursion, Tahoe Tx, and NewLimit are already working on their own models to do so. These efforts to make it accessible and easy to triage for new targets will help facilitate therapeutic abundance, breaking Eroom’s Law and bringing more drugs to patients desperately in need.
Mapping the off-target effects of every FDA-approved drug in existence (EvE Bio) [Abhishaike Mahajan, Owl Posting, July 2025]
Traditional pharma have little financial incentive to study and trial drugs once they work for their primary indications. Yet, by some measures, roughly a third of FDA‑approved drugs eventually gain a second life treating conditions for which they were not originally intended – some examples that come to mind include Gabapentin, Lenalidomide, and Minoxidil but most of this repurposing is, at-least initially, anecdotally driven and under-researched.
Now, a new Focused Research Organization (FRO), EvE Bio, is curating data on off-target effects for FDA-approved drugs. By systematically measuring both agonist and antagonist activity EvE is providing a clean, quantitative starting grid for anyone looking for new indications. Their screens are run in on harmonized assays with negative as well as positive controls, meaning there is less noise and fewer hidden biases. So far they have profiled 1,397 clinically relevant small‑molecule drugs against dozens of the body’s most druggable receptors – primarily G‑protein–coupled and nuclear receptors – producing a large map of drug–target/off‑target interactions. Their data-sets have future indications for safer prescribing, building smarter ML for biology models, and finding unexpected repurposing opportunities.
Papers
ADIOS: Antibody Development via Opponent Shaping [Towers et al., arXiv, June 2025]
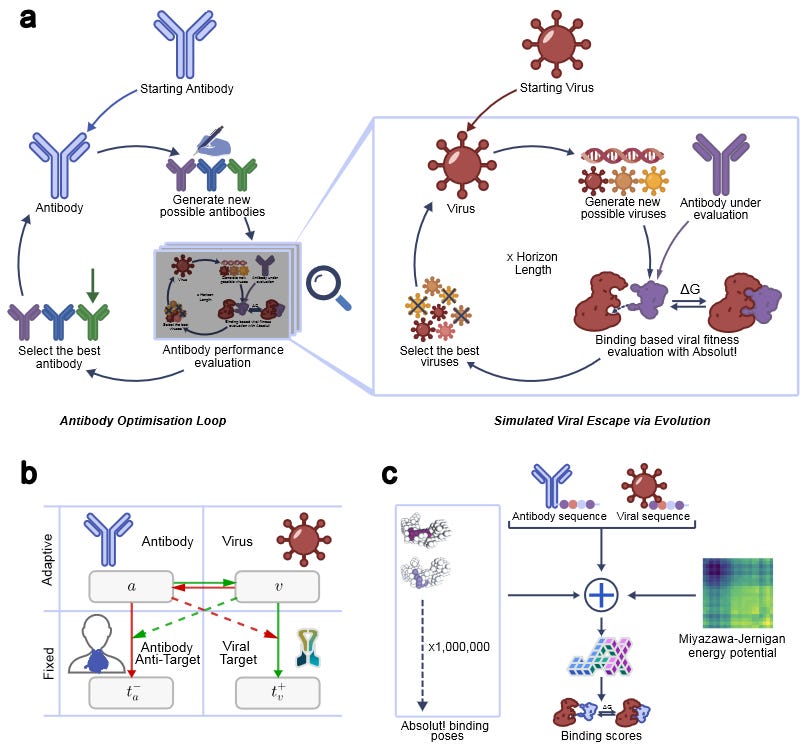
A crucial challenge in developing effective antiviral drugs is ensuring that the therapeutic modality remains effective against downstream viral variants. Indeed, the COVID-19 pandemic demonstrated the necessity for multiple booster vaccines that could maintain an immune response against mutated variants and keep the pandemic at bay. The ability to design a single therapeutic that could maintain potency against a virus without the need for updates would be a significant achievement in the overall socioeconomic cost of antiviral drugs. To that end, Towers et al. have proposed the ADIOS framework that designs antibodies that can not only adapt to viral variants, but also steer the viral escape trajectory of a virus towards less pathogenic variants.
ADIOS uses ideas from reinforcement learning, specifically opponent shaping, to mimic multi-agent interactions between an antibody and virus. The framework treats antibody-virus binding as a zero-sum game: an antibody’s reward or payoff is increased by greater strength to a virus and weaker binding to human proteins while the virus’s payoff is increased by weaker interactions with an antibody and stronger interactions with host cell receptors. In order to simulate the evolutionary pressure and viral escape, the authors use a two-layered iterative approach. In the first layer, ADIOS explores the space of viral mutations that can decrease antibody binding strength. In the second layer, the model then optimizes the initial antibody design to regain potency against the evolved mutated viruses. Noting the accuracy-speed tradeoff for methods like molecular dynamics and sequence models to predict binding, the authors developed a GPU-optimized version of Absolut! binding simulator that calculates the energy of various binding poses, and uses the binding energies to score interactions. Using their two layered approach and optimized binding simulator, the authors tested ADIOS on its ability to design antibodies for different viruses across a range of time horizons.
Comparing the shaper antibodies from ADIOS and their myopic (not meant to address to viral evolution) counterparts, the authors found the fitness of shaper antibodies significantly outpaced that of myopic candidates across dengue, West Nile virus, influenza, and MERS-CoV. Interestingly, it was found that while myopic antibodies could more effectively lower viral fitness than shapers on short time scales, shapers were more effective across longer time horizons of viral evolution. The authors concluded by noting that improvements to ADIOS could come from more accurate models of protein interactions and viral escape simulations, and discussed the possibility of integration with tools like AlphaFold3. While the most reliable test of ADIOS’s capability will be through wet lab validations, the framework proposed is a fascinating approach to designing more effective therapeutics at an overall lower cost.
AUTOCT: Automating Interpretable Clinical Trial Prediction with LLM [Liu et al., arXiv, June 2025]
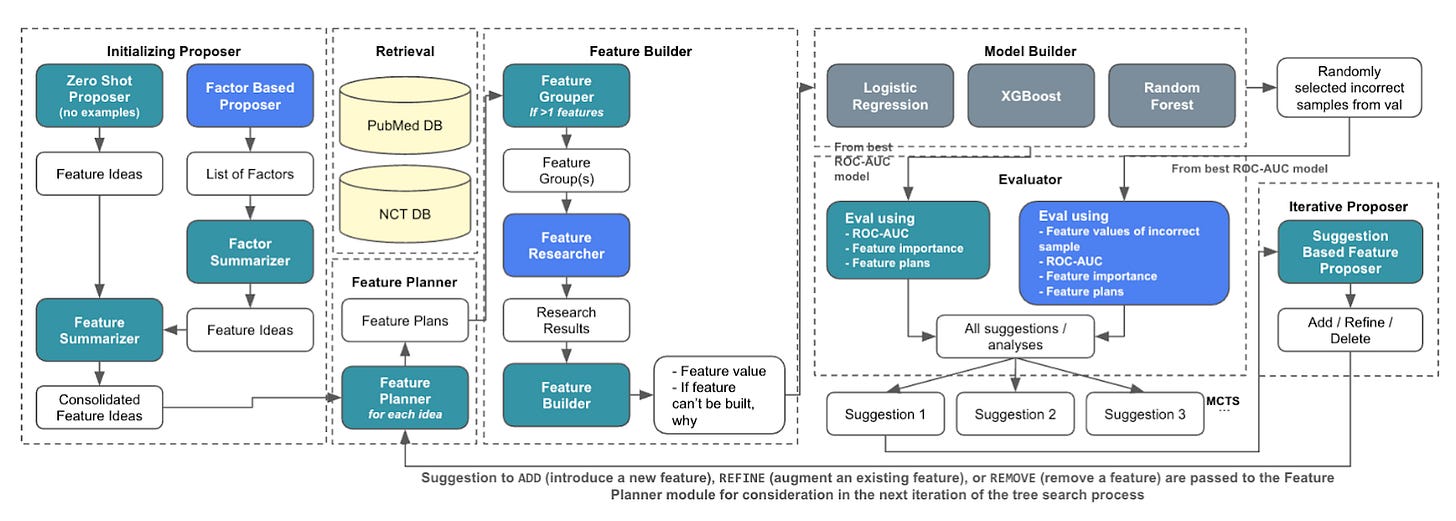
The ability to predict clinical trial outcomes could have far reaching implications to the process and cost of bringing new drugs to market. Early attempts (2016-2021) applying classical machine learning to predict trial outcomes using expert-curated features achieved robust performance but were limited by their reliance on manually annotated tabular data so couldn’t incorporate unstructured biomedical data.
Recent work (2022-2024) employing deep learning integrates diverse information such as “disease hierarchies, similarities with prior trials, drug toxicity profiles, and trial design attributes”. However, due to the ‘black box’ nature of deep learning, the predictions are hard to interpret; when it comes to decision making in clinical trials, where each decision is high stakes, interpretability and uncertainty quantification are critical.
Liu et al. introduce AutoCT (Automated Interpretable Clinical Trial Prediction with LLM Agents), which combines LLMs with classical machine learning to overcome the aforementioned limitations. AutoCT emulates the expert workflow by using LLMs for feature extraction and building (e.g. relevant trial histories, drug properties, or disease mechanisms). Subsequently, the Model Builder builds three classical machine learning models (Logistic Regression, Random Forest and XGBoost). Monte Carlo Tree Search (MCTS) is then used to explore subsequent iterations of the AutoCT pipeline to boost performance. Limited by a maximum of 10 rollouts and a depth of 10, the framework achieves an ROC-AUC of 0.753, 0.639 and 0.702 on the test set for Phase I, II and III respectively; which are comparable to deep learning approaches (HINT, SPOT and TrialBench).
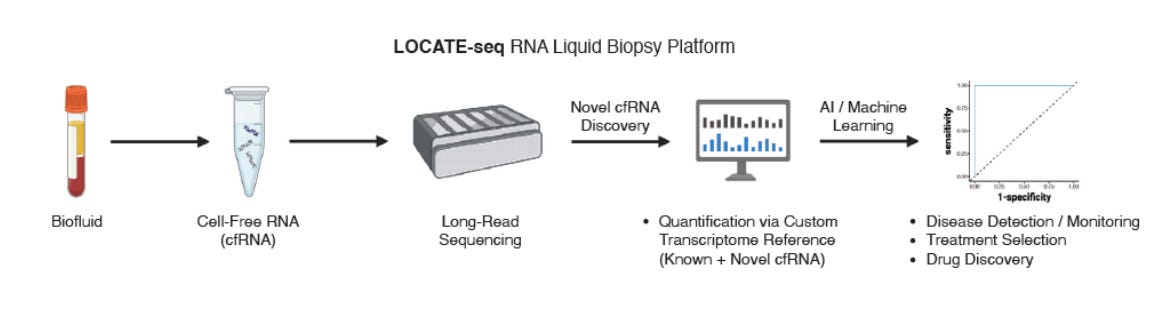
RNA liquid biopsy via nanopore sequencing for novel biomarker discovery and cancer early detection [Peddu et al., bioRxiv, July 2025]
Liquid biopsies are transforming cancer diagnostics by replacing invasive tissue biopsies with a simple blood draw. Early efforts and now commercial products such as those from GRAIL centered on detecting circulating-tumor DNA (ctDNA), but ctDNA often appears late in tumor evolution and at low abundance. New works such as those from Hani Goodarzi’s lab at UCSF have expanded the technology towards detecting circulating RNAs, which have the potential to reveal malignancy sooner because RNA is shed continuously by living tumor cells. Building on this momentum, Daniel Kim’s group at UC Santa Cruz developed LOCATE-seq, a long-read based liquid biopsy technology which uncovers long RNA biomarkers for cancer detection.
LOCATE-seq utilizes Oxford Nanopore’s long-read sequencing technology to read out RNA transcripts. Cancer is generally a disease of genomic instability - one common consequence is the dysregulation of transcription and translation. Long RNAs can form from the aberrant stitching together of RNAs during transcription or from genetic events recombining genes, which underpin famous cancers such as the canonical BCR–ABL fusion oncogene that drives chronic myeloid leukemia (CML). The team analyzed transcriptomes from 47 patients (16 healthy + 12 precancerous Barrett’s esophagus with high-grade dysplasia + 19 esophageal adenocarcinoma) using the LOCATE-seq pipeline, and discovered 276,346 previously unannotated cfRNA transcripts, 270,679 of which are in intergenic regions.
In comparing the healthy patients and diseased patients, the investigators observed a dramatic enrichment of mitochondrial cfRNAs—particularly oxidative-phosphorylation mRNAs—in plasma from both high-grade dysplasia and early esophageal adenocarcinoma. Coupling these known mitochondrial genes with the 20 most consistently up-regulated novel cfRNAs, they trained a machine-learning model that separated precancerous or stage-I cancers from controls with perfect internal cross-validation performance (100 % sensitivity and 100 % specificity). Beyond diagnostics, pathway analysis of the dysregulated transcripts flagged metabolic stress and PI3K–AKT–mTOR signalling as early vulnerabilities.
Notable deals
Merck agrees to acquire Verona Pharma for $10B. In doing so, Merck will obtain rights to Verona’s Ohtuvayre, a newly-approved treatment for COPD administered via inhalation, which analysts are projecting to potentially attain over $3B in revenue. This acquisition marks Merck’s first of the year and indicates a move to diversify their existing drug portfolio and build their presence in respiratory therapies, particularly as Merck’s cancer blockbuster, Keytruda, nears patent expiration.
After four years of development, Flagship Pioneering launches Terrana Biosciences with a $50 million initial investment. The company is creating agricultural products using RNA-based technology to improve crop resilience and protect yields without changing a plant's genetic code. By tapping into a plant's natural communication system, Terrana's platform promises to deliver targeted, temporary instructions to help crops fight disease, pests, and adapt to climate challenges. This approach is designed to give farmers more flexible and responsive tools that can be developed faster than conventional methods and applied at any point during the growing season.
Bay Area-based Centivax raises in $45 million for its unique take on a universal flu shot. An alternative to the traditional flu shot, Centivax has developed an mRNA vaccine that includes protein-encoding molecules from 22 separate flu strains—originating as far back as the Spanish flu of 1918 up to the present—in hopes of identifying a shared weak spot of the viruses that could be utilized to encourage generalizable immunity. Animal studies have thus far shown promise, and based on Phase 2 results in human subjects, Centivax hints at potentially pursuing accelerated approval for 2029. The round was led by Future Ventures with participation from NFX, BOLD Capital Partners, Base4 Capital, Kendall Capital Partners, and AmplifyBio.
Novartis secures the option to acquire Sironax’s blood-brain-barrier delivery platform for various therapeutic modalities at a price of up to $175M in upfront and near-term payments.
ScienceMachine has successfully raised $3.5 million in a pre-seed funding round, co-led by Revent and Nucleus Capital. This investment will fuel the development of ScienceMachine’s autonomous AI agents which seek to expedite scientific discovery by significantly lowering time, hiring, and cost constraints for pharma and biotech companies. Their existing agent and flagship product, Sam, accomplishes this by functioning as an innovative "always-on" AI bioinformatician designed to seamlessly integrate with existing databases, workflows, and models. Sam continuously processes data to perform advanced analysis, uncovering hidden insights and patterns that can accelerate scientific discovery.
In case you missed it
Biomni accelerates biomedical discoveries by 100x with Claude
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.