BioByte 121: biology as the next software platform, language development in children, a massive Perturb-seq dataset, and morning coffee as a cancer treatment
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
Reminder to sign up to join us for a reception to close out our third annual AI x Bio Summit on July 22! Enjoy an evening of wine and conversation beneath the lights of the storied New York Stock Exchange trading floor as we reflect, connect, and celebrate. It’s a chance to engage with fellow founders, researchers, and operators shaping the future of biology and technology. Spaces are filling up fast so please register to reserve your spot here. We can’t wait to see you there!
What we read
Blogs
Why Biology Is the Next Software Platform [Ryan Roddy, Ryan Roddy, June 2025]
Technological advances accepted to be defining moments in history—like the advent of the internet or the moon landing—are not simply accomplishments reached in isolation but rather the culmination of countless efforts within a climate conducive to their success. This belief is readily evident when considering the AI revolution, a paradigm shift largely facilitated by decreasing cost and increasing accessibility of compute. Ryan Roddy, an MP at Seaside Ventures, postulates that the current climate in 2025 surrounding programmable biology postures the field to undertake its own defining moment. He likens the state of biology to software, pointing to the technological advances made in reading and writing biology as well as several cost curves—such as cheaper DNA sequencing and lab automation—that will help enable creative thinkers to rapidly iterate, greatly shortening experimental turnaround time and increasing output. Additionally, Roddy points to tools like protein structure prediction via AlphaFold, resources like commercially available gene fragments, political tailwinds, and high capital availability as other factors that are facilitating this transformation of biology into a field that is iterative, modular, fast, and cheap.
Software-like, programmable biology will enable the development of transformative solutions to challenging problems. Crops and plants could be easily edited to bolster resistance to rapid environmental changes. Microbes could be engineered to produce large volumes of otherwise difficult to synthesize molecules, while gene therapies could become commonplace. However, there are risks and limitations inherently associated with advanced biological tools that cannot be ignored. Biosafety is of particular import, as rogue agents could maliciously leverage these capabilities to destroy ecosystems. Furthermore, biology is still a physical science, so scaling biological solutions to commercial viability necessitates significant CapEx and infrastructure investments. Despite these shortcomings, decreasing costs of fundamental tools and resources have historically led to groundbreaking advances. Who’s to say that biology will be any different?
Papers
Emergence of Language in the Developing Brain [Evanson et al., Meta AI, May 2025]
A child’s brain has a unique ability to acquire language, but when and how the neural representations that underpin such acquisition occur remain poorly understood. To study the neural representations of language across development, the team implanted 7,427 intracranial electrodes in the brains of 46 children, teenagers and adults as part of their treatment of epilepsy (via stereotactic EEG electrodes). The participants then listened to an audiobook recording of Le Petit Prince and the elicited brain signals were analyzed.
Using encoding and decoding models targeting features that were predicted by theoretical linguistics or learned by LLMs, the authors map the emergence, timing and localization of language representations across the cortex.
They demonstrated that a large set of language features is already present in the cortex of 2-5 year olds, with neural dynamics similar to adults but restricted to a narrow set of regions that gradually expand during development. These anatomical-functional changes are accompanied with a development of the hierarchy of language representations.
The authors also draw parallels with how AI models learn language representation by observing two AI models (wav2vec 2.0 and Llama 3.1; trained on speech and text):
Both models learn representations that are similar to all three groups
Wav2vec 2.0 develops a layered structure that mirrors the anatomical and temporal hierarchy of language processing in the brain, which is not observed in Llama 3.1
Training the models strengthens the similarity with the developing brain
The representations acquired by Llama 3.1 through training can only be observed in the brain of the oldest individuals
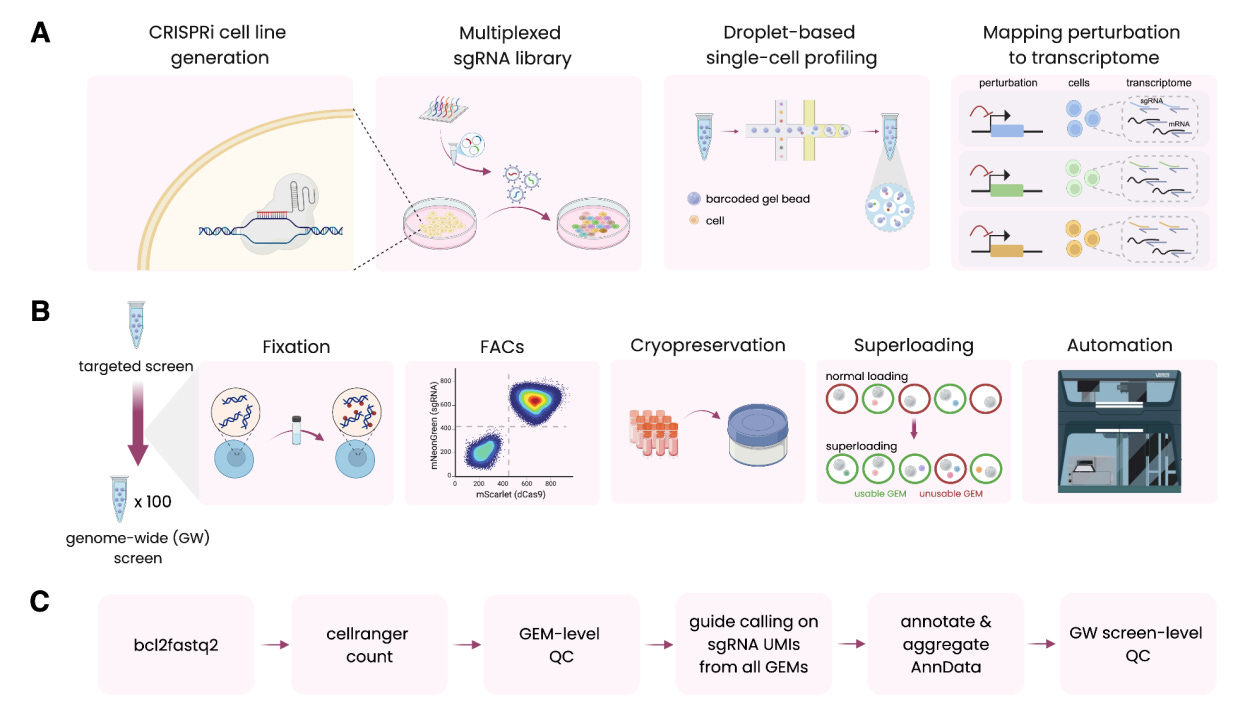
X-Atlas/Orion: Genome-wide Perturb-seq Datasets via a Scalable Fix-Cryopreserve Platform for Training Dose-Dependent Biological Foundation Models [Huang et al., bioRvix, June 2025]
Recently, there has been a marked increase in the development of foundation models for biology across a variety of domains. Notable examples include the Evo series of genome language models from the Arc Institute and the Boltz-2 protein structure and binding affinity model from MIT and Recursion Pharmaceuticals. However, a current limitation of foundation models is that they struggle to identify causal relationships in data, specifically the mechanisms by which perturbations may affect a cell’s disease state. Of course, such models require reams of high-quality training data, usually from Perturb-seq experiments that quantify the effect of modifications to a cell’s genetic state on downstream phenotypic outcomes.
Garnering over one billion dollars in startup investment, Xaira Therapeutics launched with great fanfare in April 2024, outlining a vision for drug discovery centered around machine learning research and novel data generation at scale to enable model development. One year after their inception, the company has debuted their Fix-Cryopreserve-ScRNAseq (FiCS) Perturb-seq pipeline and a massive dataset named X-Atlas/Orion (Orion referring to the version name). Noting that “current foundation models often struggle to robustly infer causal relationships and predict the outcome of specific interventions” in their preprint, scientists at Xaira point to the need for large scale Perturb-seq datasets to drive the development of “virtual cell” models that can better represent the effects of different drugs on disease states.
The authors first identify primary limitations of conventional Perturb-seq datasets, specifically a dependence on freshly harvested cells, the timescale bottleneck of fluorescence-based sorting, and the associated quality control and automation requirements to scale to larger throughput studies. The newer FiCS platform uses chemical preservation techniques immediately after sample harvesting to minimize induced cellular stress that may influence final results. Furthermore, the platform makes use of optimized staining methods to improve sorting and automates library preparation with liquid handler robots to reduce variation across batches. Beyond the improvements in methodology itself, the authors validate the FiCS pipeline by measuring perturbations of every human protein-coding gene (nearly nineteen thousand) in HCT116 and HEK293T cell lines. After ensuring that the results of the FiCS pipeline were consistent with established benchmarks of Perturb-seq performance, the team trained linear classifiers to identify actually perturbed cells against control populations. The team also compared the correlations of gene interactions identified from their FiCS atlas with benchmarks from the StringDB database, concluding that their methods could recapitulate known gene-gene dependencies by measuring the effects of targeted perturbations.
The paper also investigated factors that influence the efficacy of gene knockdown (KD). Noting that the two cell lines showed differing levels of gene inhibition despite using identical protocols and delivery systems, the authors attributed the difference to varying levels of sgRNAs and scaffolding protein factors like TRIM28. Such results are useful features to assess the performance of the CRISPRi system that actually makes the perturbations and to control for downstream analysis. Finally, it was shown that there is a high correlation between knockdown efficiency and the level of sgRNA in cells. Such measurements are useful for trying to measure dosage-dependent effects of perturbations. Such inference is made difficult when solely using scRNA-seq data due to artifacts from the actual data generation process like high sparsity and transcript dropout. The ability to use sgRNA abundance data as a proxy for KD efficiency represents a promising alternative to better understand a range of perturbation effects.
Humanized Caffeine-Inducible Systems for Controlling Cellular Functions [Scheller et al., bioRxiv, June 2025] / Caffeine-regulated molecular switches for functional control of CAR T cells in vivo [Sylvander et al., bioRxiv, June 2025]
What if your morning cup of coffee could also fight your cancer?
In 2006, Ladenson et al. immunized a llama with caffeine, producing a caffeine-neutralizing nanobody. Subsequent kinetic studies by Sonneson & Horn in 2009 revealed that this nanobody dimerizes around caffeine rather than binding it in a 1:1 ratio. This finding unlocked new possibilities in synthetic biology, as careful engineering could exploit caffeine to recruit and dimerize any protein of interest tagged with this nanobody. This caffeine-nanobody system presents intriguing prospects as an inducible therapeutic platform, given caffeine's affordability, safety, and ability to cross the blood-brain barrier. Recently, parallel studies by Scheller et al. and Sylvander et al. expanded this technology, meticulously adjusting caffeine sensitivity and demonstrating its practical application for controlling CAR-T cell activity in treating cancer.
Initially, the original nanobody formed a homodimer (A-A). Both groups improved the design by engineering two distinct nanobodies ('B', 'C'), enabling heterodimerization (B-C). This heterodimerization provided two key benefits: first, it allowed recruitment of two separate halves of split proteins without the requirement for homomeric interactions; second, it enabled more precise control of caffeine sensitivity by separately engineering the monomers. Scheller et al. utilized a physics-based computational pipeline (varCOMETS) for initial designs, whereas Sylvander et al. employed yeast display techniques to optimize their nanobodies. Both groups selected heterodimers with a 5–10-fold reduction in caffeine sensitivity compared to the original nanobody, reasoning that less sensitivity would decrease background activation, as higher caffeine doses are tolerable in humans. Scheller et al. also carefully crafted some extra mutations to reduce the immunogenicity of the llama-derived nanobody.
Both groups tested their engineered caffeine-responsive systems for controlling CAR-T cell functionality. Scheller et al. implemented a custom caffeine-inducible transcription factor that drove HER2 CAR-T expression. Sylvander et al. used the caffeine-inducible system to dimerize CAR receptors themselves - with one monomer containing the CD19 binding domain while the other CAR monomer contained the CD3ζ signaling domain necessary for CAR activation. Both groups demonstrated successful CAR expression and activation, but Sylvander et al. significantly expanded the system’s in vivo validation, demonstrating tumor clearance in mouse models. Importantly, Sylvander et al. also showed that intermittent caffeine administration reduced CAR-T cell exhaustion and mitigated the hyperactivity observed with CAR constructs containing overactive cJun, addressing a challenge previously linked to a phase 1 clinical trial failure.
A computational approach to evaluate how molecular mechanisms impact large scale brain activity [Sacha et al., Nature Computational Science, May 2025]
Understanding how molecular-level changes shape whole-brain activity is a central challenge in neuroscience, especially for developing better treatments that affect consciousness, like anesthetics or psychiatric drugs. While many of these compounds act on specific synaptic receptors, their effects are typically measured using large-scale brain imaging techniques such as EEG or fMRI. To link these very different scales, Sacha and colleagues developed a multiscale modeling approach. They started with detailed neuron and synapse models and used them to build simplified representations of neural populations, known as mean-field models. These were then placed into a brain-wide network based on real anatomical data to simulate full-brain activity. By adjusting synaptic parameters to mimic the action of drugs like propofol and ketamine, the model was able to reproduce patterns seen during unconscious states, such as widespread slow-wave oscillations and reduced responsiveness to external input. The simulations also showed a stronger match between structural wiring and functional activity under anesthesia and sleep.
Notable deals
Eli Lilly is acquiring Verve Therapeutics for up to $1.3 billion to strengthen its position in genetic medicines for cardiovascular disease. Verve is developing single-dose gene editing treatments aimed at lifelong reduction of cardiovascular risk, including its lead candidate VERVE-102, which targets the PCSK9 gene and is currently in Phase 1b trials. The deal aligns with Lilly’s expertise in cardiometabolic care and is expected to close in Q3 2025.
Draig Therapeutics launches with a $140M Series A to treat depression. The UK-based biotech is pushing its lead asset DT-101 through phase II trials this year. DT-101 balances the excitatory-inhibitory activity throughout the brain by focused on a brain receptor called AMPA. The round was led by Sanofi Ventures, Canaan and Access Biotechnology.
Supernus Pharmaceuticals will be acquiring Sage Therapeutics for up to $795M. With this move, Supernus seeks to bolster their CNS portfolio, diversify their revenue base, and increase their cash flow—objectives the acquisition of zuranolone, the only approved oral medicine for postpartum depression, will help them achieve. Five months earlier, Sage had turned down an acquisition offer of $469M from Biogen, believing it to undervalue the company.
In case you missed it
FDA to Issue New Commissioner’s National Priority Vouchers to Companies Supporting U.S. National Interests | FDA
The FDA announces the Commissioner’s National Priority Voucher, a new structure for reviewing drug admissions. The turnaround time for FDA approvals currently sits at 10-12 months; they aim to reduce this to 1-2 months. The process centralizes experts for a team-based review, in contrast with the current process which sends out review requests to separate FDA sites. The first year, they will only give vouchers to companies with “U.S. national priorities”.
What we listened to
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.