BioByte 120: FutureHouse launches ether0, introducing Origene as a virtual disease biologist, newly released Boltz-2 targets binding affinity, and combating negativity besieging biotech
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
Reminder to sign up to join us for a reception to close out our third annual AI x Bio Summit on July 22! Enjoy an evening of wine and conversation beneath the lights of the storied New York Stock Exchange trading floor as we reflect, connect, and celebrate. It’s a chance to engage with fellow founders, researchers, and operators shaping the future of biology and technology. Spaces are filling up fast so please register to reserve your spot here. We can’t wait to see you there!
What we read
Blogs
“We Got This”: Don’t Let Negativity Become a Self-fulfilling Prophecy [Arthur Tzianabos, LifeSciVC, June 2025]
With so many negative headlines deluging the biotech and pharma industries as of late—valuation decreases and down rounds, governmental funding cuts, reductions in IPOs, layoffs, and broader regulatory and economic challenges—it’s easy to feel defeated in the current environment. CEO of Lifordi Immunotherapeutics, Arthur Tzianabos, however, writes to the biotech community with the important reminder that we must not succumb to this cynical narrative. In doing so, we risk perpetuating pessimism, further manifesting such fatalistic attitudes into reality like a self-fulfilling prophecy. Certainly this is easier said than done, so how can we fight back against all the gloom and cynicism?
Tzianabos answers this proposition with many vital reminders which contextualize and support his emphatic rallying cry. He splits these facts into four categories which he cites as the key ingredients to the biotech recipe:
Solid science that produces promising drug candidates: it is important to note that critical scientific discoveries are still being made as transformative drugs continue to successfully reach the market or are rapidly barreling towards approval, such as various CAR T-cell therapies, GLP-1 receptor agonists, and PD-1 inhibitors.
Well-weathered leadership: as has been seen in the past, difficult times create stronger and more agile leaders. Those in upper-management roles today have weathered several challenging biotech/pharma eras, learning to be nimble and open-minded throughout.
Consumer demand: Even amidst the downturn, clinical trials and clinical trial recruitments remain high. Tzianabos cites that there are currently 20,465 ongoing trials in the US and 65,474 recruiting globally.
Access to funding: Perhaps most uplifting is the state of available capital and funding. Investors have dry powder to deploy—data from Pitchbook and Cambridge Associates estimates a whopping $1.7 trillion of undeployed capital. Additionally, around $340 billion raised in 2019 to 2021 is nearing the end of its investment period, meaning it will have to be deployed in the near future. Moreover, pharma companies continue to shop around for acquisitions, especially as some key drugs face patent expirations. In 2025 alone, M&A in the life sciences accounted for $30 billion of announced value.
In addition to these trends, the author contextualizes the cause for much of the negative news in the industry. During the biotech boom, many companies with bloated valuations elected to IPO at far earlier stages than usual, oftentimes even while they were preclinical. Now whilst they try to bring these drugs through the early stages of clinical trials, they are subject to the unforgiving judgment of the public markets—a process usually confined to the private stages. These early readouts are also often very open to interpretation or meaningless, so placing them under public scrutiny in an environment with a pessimistic narrative lends itself to further negativity. Finally, many treatments like the Covid-19 vaccines were fast-tracked during the pandemic, leading to unrealistic expectations of drug development timelines. Some of the frustration felt today is simply a renormalization to the lengthy process of developing and approving drugs after the unique conditions of the Covid-19 pandemic.
Tzianabos ends by urging the industry to break the narrative, embrace a positive mindset, and continue doing great work to bring about transformative drugs for patients in need. We’ve got this!
The 23andMe Deal [Cremieux, May 2025]
With the acquisition of 23andMe, Regeneron has acquired the ability to prioritize targets to increase the odds of success of the drugs they pick, do target discovery better than anyone is capable of doing, understand side effects profiles, reliably do repurposing and treat side effects pharmacologically. The whole article is worth a detailed read, but we’ll discuss target prioritization below.
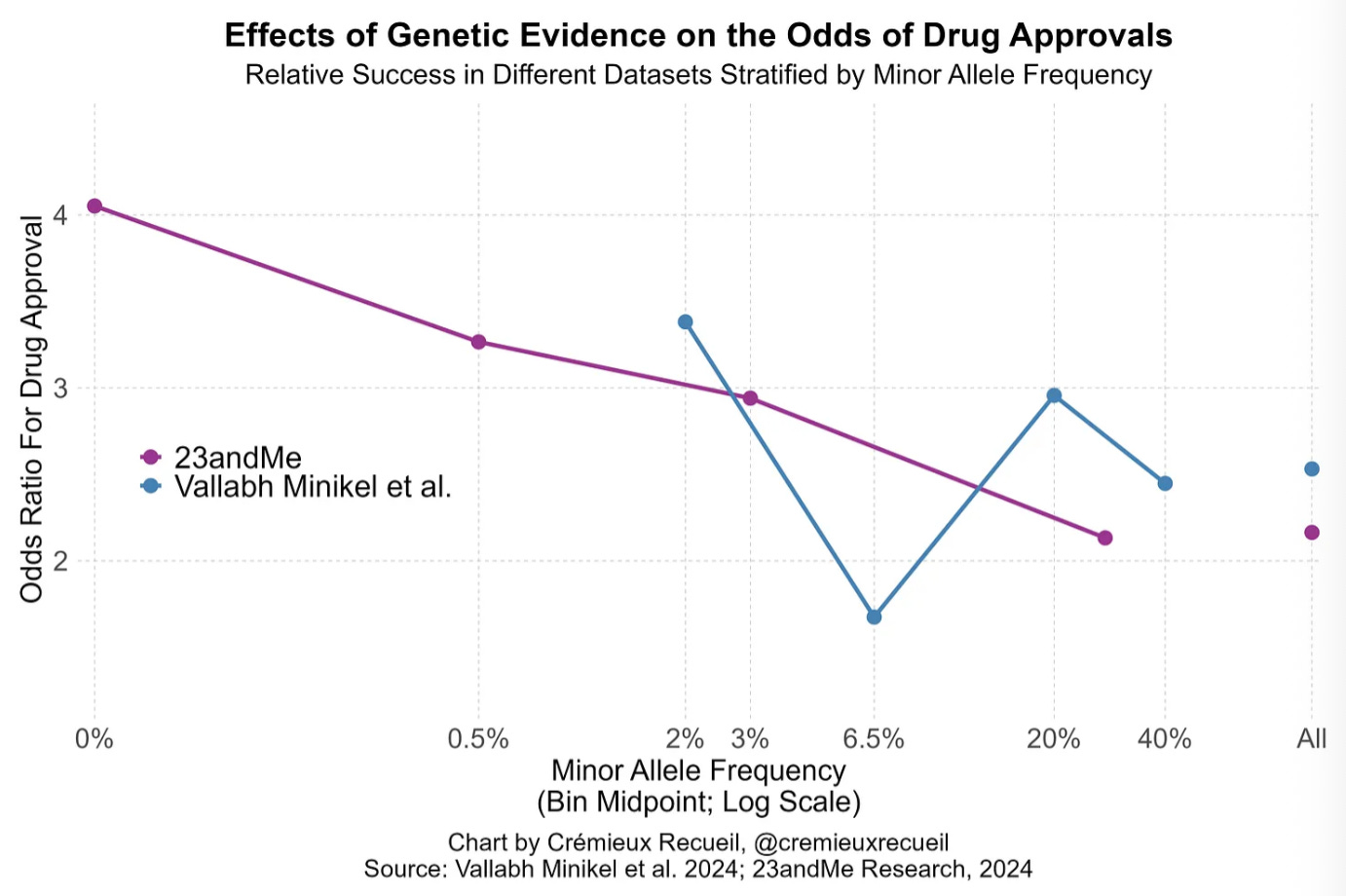
In a groundbreaking 2015 study, Matthew Nelson et al. demonstrated that drugs targeting mechanisms supported by genetic evidence had double the odds of regulatory approval. A 2019 follow-up study with more data (King et al.), found that genetic support was even more strongly related to a likelihood of approval; especially if the evidence was related to Mendelian traits and GWAS associations were linked to coding variants. In 2024, a new study by Minikel et al., found that genetic evidence was associated 2.6x higher odds of approval.
The author highlights that this last study had a major weakness. The data they accessed did not provide them with huge statistical power to detect “the positive association between the effect sizes of different variants or the inverse of their minor allele frequencies and the odds of clinical success”. In a subsequent paper, 23andMe was able to demonstrate that both of these factors influenced drug approval odds. The 23andMe team demonstrated that because of its larger database size, their results were better predicted by theory (see graph above). Because of this, their dataset is more likely to predict success. They were also able to prove a better indication if variants were causal.
Papers
OriGene: A Self-Evolving Virtual Disease Biologist Automating Therapeutic Target Discovery [Zhang et al., bioRxiv, June 2025]
Scientists in China have developed OriGene, a “virtual disease biologist” that doesn’t just read papers—it reasons across 568 biology-specific tools, proposes new drug targets, and even designs the wet-lab assays needed to validate them.
Under the hood, five agents—Coordinator, Planning, Reasoning, Critic, and Reporting—pass ideas around in a tight feedback loop that mimics how a real scientist brainstorms, checks, and refines a theory. Each agent can call on 568 specialized databases and analytical tools spanning disease biology, pharmacology, and competitive‑landscape intel, all exposed through the OriGeneHub API. Letting that loop “think harder” demonstrably helps: OriGene’s LitQA accuracy climbs from 62.81% with a single sub-query (1× budget) to 78.39% when the Coordinator allows nine iterations (9×). The system also harvests successful reasoning trajectories into “thinking templates” that guide future Planning and Coordinator steps; however, the authors only claim qualitative gains from this Level‑2 template evolution and do not report hard numbers to prove it.
To test whether all that machinery actually helps, the team built TRQA, a 1,921‑question benchmark split into two halves. TRQA‑lit mines the literature (172 multiple‑choice + 1,108 short‑answer items), while TRQA‑db pulls 641 short‑answer questions from commercial drug‑pipeline databases. Importantly, any “too‑easy” questions that a baseline LLM (Doubao without search) could already answer were thrown out, ensuring the remaining items demand non‑trivial reasoning and retrieval. On these tougher sets OriGene scores 60.1 % accuracy on TRQA‑lit multiple choice, 82.8 % recall on TRQA‑lit short answer, and 72.1 % recall on TRQA‑db—comfortably ahead of human experts with Google and ahead of GPT‑4o‑search or TxAgent.
As any good comp-bio-powered biology paper should, the authors put OriGene to the test on two hard cancers. In hepatocellular carcinoma, the agents started with 125 candidates and converged on GPR160, spotting its up‑regulation in tumor datasets and an inverse link to recurrence‑free survival (p = 0.0057). It then designed a suite of assays—gene knock-down, cytokine profiling, and immune-context cocultures—to probe GPR160’s role and specifically measure how inhibition boosts CD4⁺/CD8⁺ T-cell infiltration in patient-derived organoids and tumor fragments. A small molecule inhibitor was developed for GPR160 (albeit seemingly not through OriGene), ultimately demonstrating significant anti-tumor activity in patient-derived organoid and tumor-fragment models mirroring human clinical exposures. A parallel run in colorectal cancer nominated ARG2; follow‑up assays showed a dose‑dependent kill curve with an IC₅₀ of 3.09 µM in HCT116 cells and significant viability drops across four patient‑derived organoids.
Boltz-2: Towards Accurate and Efficient Binding Affinity Prediction [Passaro et al., preprint, June 2025]
Late last week, the team of MIT scientists behind Boltz-1, now partnered with Recursion Pharmaceuticals, released the second iteration of the Boltz series to much fanfare within the community.
The release of AlphaFold3 by Google DeepMind in May 2024 ushered in significant advancements in biological structure prediction, building on its predecessor models to allow prediction of protein complexes with nucleic acids and small molecules. Open-source versions soon followed, with the Boltz-1 model being the first to achieve accuracy comparable to AlphaFold3 and being widely available for both academic and commercial purposes. Apart from general improvements in structure prediction metrics, Boltz-2 also builds upon its predecessor by allowing users to impose conditions during inference, like distance constraints between residues. The authors also added various datasets of short molecular dynamics trajectories to their training set, allowing Boltz-2 to predict local protein dynamics and conformational ensembles.
Boltz-2’s most important contribution is its binding affinity prediction modules. Characterizing the binding affinity of targets is a key bottleneck step in the drug development cycle; despite older models offering high-fidelity structures, they are unable to predict the physical binding affinity of proteins and ligands, leaving researchers stuck with computationally expensive physics-based methods like free-energy perturbation (FEP) simulations or relatively inaccurate docking tools.To that end, the authors evaluate Boltz-2 on its ability to predict not only whether or not a molecule is a real binder for a given target, but also with what physically measurable affinity it would bind. For binding affinity, Boltz-2 showed high correlation with the results of benchmarks for both hit-to-lead and lead-optimization tasks. Notably, it delivered that performance comparable to FEP simulations, while only requiring a fraction of the time to produce results. The authors admit that Boltz-2 did have some weaknesses, noting that the model struggled to show appreciable correlation when tested on unseen internal assays provided by Recursion. Boltz-2 was also used to perform a virtual screen for the well-studied TYK2 target. Screening over 500,000 compounds from candidate libraries, Boltz-2 successfully identified the majority of top known compounds as binders, while crucially also identifying non-binders with high accuracy. It is worth noting that the binding affinity prediction task required a significant data engineering effort, specifically in order to standardize the range of reported measurements for affinity based on the assays used to generate them. Due to the variances in assays and the equations used to measure affinities, the model’s predictions are called “IC50-like” and meant to be used more for relative ranking.
In summary, the addition of downstream modules for binding affinity prediction shows the immense potential of structural foundation models like Boltz-2 for various tasks. Additionally, the integration of molecular dynamics data and user-defined constraints should hopefully allow easier adoption over a greater range of research (on the day of its release, Boltz-2 was made available on a multitude of online drug design and simulation platforms). However, Boltz-2 does have some limitations. The model still lags behind AlphaFold3 on structural benchmarks, most visibly on antibody tasks. Additionally, while the model showed comparable performance with other protein conformation models like AlphaFlow and BioEmu, all three models were trained on relatively short MD trajectories (mostly ~100ns), and it remains to be seen whether they can scale to protein dynamics that can occur over longer timescales. Nonetheless, the speed of Boltz-2’s adoption, including being now offered on major platforms like NVIDIA’s BioNeMo, should allow for a more comprehensive picture of the model’s capabilities in the coming months.
Training a Scientific Reasoning Model for Chemistry [Narayanan et al., preprint, June 2025]
After a slew of recent UI releases, the ever-busy FutureHouse team are back with their latest release: ether0, a model that reasons in natural language and outputs chemical structures as SMILES strings. A clear application of such a model is in molecular design. To illustrate a use case, the team show a sample problem and answer using ether. In this example, the model shows exploration, backtracking and then verification.
Latest models such as OpenAI’s o3 have shown strong performance on chemistry benchmarks that only involve natural language, however have failed when chemical structure is introduced. This is one of the drivers for the team to create a model that excels here. The team combined SOTA reasoning models with reinforcement-learning post-training with verifiable rewards, which the team carefully crafted a dataset for. One challenge for the team has been to prevent the model ‘forgetting’ its natural language capabilities as it trains using RL on the chemical structures.
Building on these motivations, ether0 is a 24-billion-parameter LLM (based on Mistral-Small-24B) that was first fine-tuned on long “chain-of-thought” traces to prime its reasoning abilities, then specialized via Group Relative Policy Optimization across seven chemistry sub-tasks (e.g., retrosynthesis, solubility editing), and finally distilled into a unified generalist model with further RL and a safety-alignment phase . In total, the team trained on 640,730 experimentally grounded problems spanning 375 tasks, yet required far less data than dedicated chemistry models.
On held-out benchmarks, ether0 not only outperforms general-purpose LLMs (Claude, o1, o3) on open-answer chemistry questions, but also beats specialized tools and even human experts in molecular design tasks. A dedicated safety-alignment phase then teaches ether0 to refuse unsafe chemistry prompts. FutureHouse is releasing the model weights, benchmark data, and reward functions to fuel further research.
Notable deals
Parallel Bio, a Y Combinator-backed startup, has raised $21 million in a Series A round led by AIX Ventures. The company is developing lab-grown human immune organs, starting with lymph nodes, to improve drug testing and development. To build these organs, Parallel Bio collects tissue samples from human donors representing various components of the immune system. These samples are combined into a cell bank the company uses to grow functional immune organs in vitro. Currently, the company partners with pharmaceutical firms to screen compounds more precisely using its organoids. Over time, it aims to compile data from these tests to train a foundation model of the human immune system. Parallel Bio also plans to eventually develop its own therapeutics, leveraging its proprietary platform and accumulated insights.
Bristol Myers Squibb has agreed to acquire PhiloChem for $350 million upfront, deepening its investment in radiopharmaceuticals. PhiloChem’s lead asset, OncoACP3, is a prostate cancer therapeutic currently in Phase I. OncoACP3 targets ACP3, offering a mechanistic alternative to Novartis’ Pluvicto, which targets PSMA. Manufacturing of the compound will be handled by RayzeBio, BMS’s prior radiopharma acquisition.
Eli Lilly has entered a licensing agreement with Swedish biotech Camurus. The deal has an undisclosed upfront and is worth $870M in biobucks all for access to its FluidCrystal delivery technology. The platform enables extended drug release via a gel that forms after injection, offering dosing that can last days to months. Lilly plans to apply the technology to up to four obesity-related compounds as it seeks to defend and extend its leadership in the GLP-1 space.
Amplify Partners has raised a $200 million fund to back technical founders building at the intersection of biology and computation. The new fund, focused on "Digital Biology," will be led by Elliot Hershberg. This vehicle complements Amplify’s existing $700 million fund, which supports portfolio companies and invests across a range of technical disciplines. The Digital Biology fund reflects a growing thesis around computation-driven innovation in life sciences.
Signify Bio Launches With an Oversubscribed $15 Million Initial Financing and Strategic Partnership With UTSW Focused on Nucleic Acid Enabled Protein Therapeutics. Other investors include Eli Lilly and Co., the Gates Foundation Strategic Investment Fund, BrightEdge, and Danaher Ventures LLC. The company—which spun out of the Siegwart Lab at the University of Texas Southwestern Medical Center—seeks to control the production, secretion, and localization of proteins for personalized therapeutics using their three platforms: Signal peptide Engineered Nucleic acid Design (SEND), iPhos, and an in-house computational platform.
In case you missed it
Crypto billionaire Brian Armstrong is ready to invest in CRISPR baby tech
Recursion to lay off 20% of staff, a month after slashing its pipeline
IQVIA and NVIDIA Harmonize to Accelerate Clinical Research and Commercialization With AI Agents
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.