BioByte 116: scaling test-time compute for antibody design, HealthBench sets the bar high for clinical LLMs, and revealing the structural secrets of human sweet receptors
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
We have two exciting announcements this week!
We’re excited to announce our collaboration with AlphaXiv! Under this new partnership, whenever we cover an arXiv paper, we’ll link to AlphaXiv, where you can read comments from other researchers and use on-site AI tools to summarize the article. This collaboration is an important first step in Decoding Bio’s mission to translate technical developments and trends into digestible formats.
We warmly invite the Decoding Bio community to join us for a reception to close out our third annual AI x Bio Summit on July 22. Enjoy an evening of wine and conversation beneath the lights of the storied New York Stock Exchange trading floor as we reflect, connect, and celebrate. It’s a chance to engage with fellow founders, researchers, and operators shaping the future of biology and technology. We can’t wait to see you there! Please register here
What we read
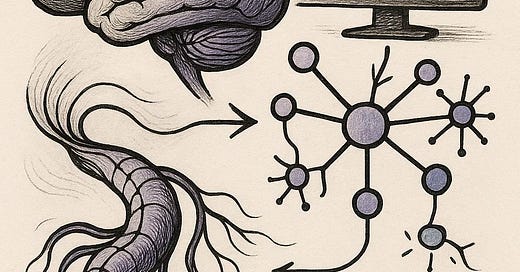
De novo design of hundreds of functional GPCR-targeting antibodies enabled by scaling test-time compute [Nabla Bio, May 2025]
Test-time compute is coming to protein design.
In the sequel of their story in designing antibody therapeutics for GPCRs, the team at Nabla Bio unveil that allowing their generative model JAM to “introspect” for longer leads to more and better binders. While they don’t reveal all the secrets of the model, at a high-level the engine under the hood is an iterative “best of N” rejection sampling algorithm: generate N proposals, filter out the best m, and use those to seed the next set of N proposals. In their first paper, they show that their generative model JAM (which co-generates antibody sequence and structure jointly) successfully designs nanomolar antibodies. They start to explore this “introspection” method, going up to three rounds of introspection. In this work, they extend this introspection up to six rounds, finding that the hit rates and affinities increasingly improve.
This level of efficiency (hit rate) and effectiveness (affinities) also leads to nonlinear functional gains. While 45 out of 47 their antibody binders targeting CXCR7 were antagonists (non-activating), they discovered that 2 (4%) were actually functional agonists (activating CXCR7). Notably, when they run this antibody agonist back through JAM to generate new functional agonists, 30% of the ~900 designed antibodies were also agonists, with half demonstrating improved production yields and half demonstrating improved monomericity.
Nabla continues to push the field forward in using generative models for protein design, revealing an exciting new scaling law to explore.
HealthBench: Evaluating Large Language Models Towards Improved Human Health [Arora et al., arXiv, May 2025]
OpenAI’s HealthBench sets a new bar for scale and diversity in health‑oriented evaluation, and sets the new bar for clinical LLMs to overcome. The benchmark assembles 5000 open‑ended conversations written by 262 practising physicians who collectively speak 49 clinical languages and have experience in 60 countries. Every conversation is paired with a physician‑authored rubric whose individual items carry positive or negative point values; in total there are 48,562 rubric points spread across five behavioural axes such as accuracy, completeness and context‑awareness, giving unusually fine‑grained leverage for scoring.
Unlike the differential‑diagnosis studies of McDuff et al. (2025) and Zhou et al. (2025)—which judge success by whether the correct label appears in a top‑k list—HealthBench asks how well a model handles many kinds of health interactions (triage, global health, data transformation, etc.). Its rubric system grades one axis at a time and then collapses to an overall score. In addition, the authors introduce a “worst‑at‑k” reliability curve that shows how quickly the poorest response quality deteriorates as more samples are taken - important in medical contexts where reducing harm is paramount. This design broadens coverage and surfaces tail‑risk behaviour that is invisible in diagnosis‑only leaderboards.
Early results reveal a distinctive performance ladder. On the full test set, o3 leads all frontier models with 0.60, but still falls to 0.32 on the intentionally difficult HealthBench‑Hard subset, leaving ample head‑room for future models. When physicians are allowed to revise earlier (Sept‑2024) model drafts their answers outrank the model alone, producing the ordering Human + AI > AI > Human; for the latest April‑2025 completions physicians can no longer add consistent lift, suggesting that collaboration benefits compress as model quality rises. This pattern contrasts with AMIE’s DDx study, where the standalone system outperformed assisted clinicians, yielding AI > Human + AI > Human.
By moving evaluation from single‑task accuracy to multi‑axis safety, completeness and worst‑case robustness, HealthBench reframes progress in medical language models from “spotting the right diagnosis” to “delivering reliable, context‑aware care at scale”—and the frontier still sits barely halfway up the rubric.
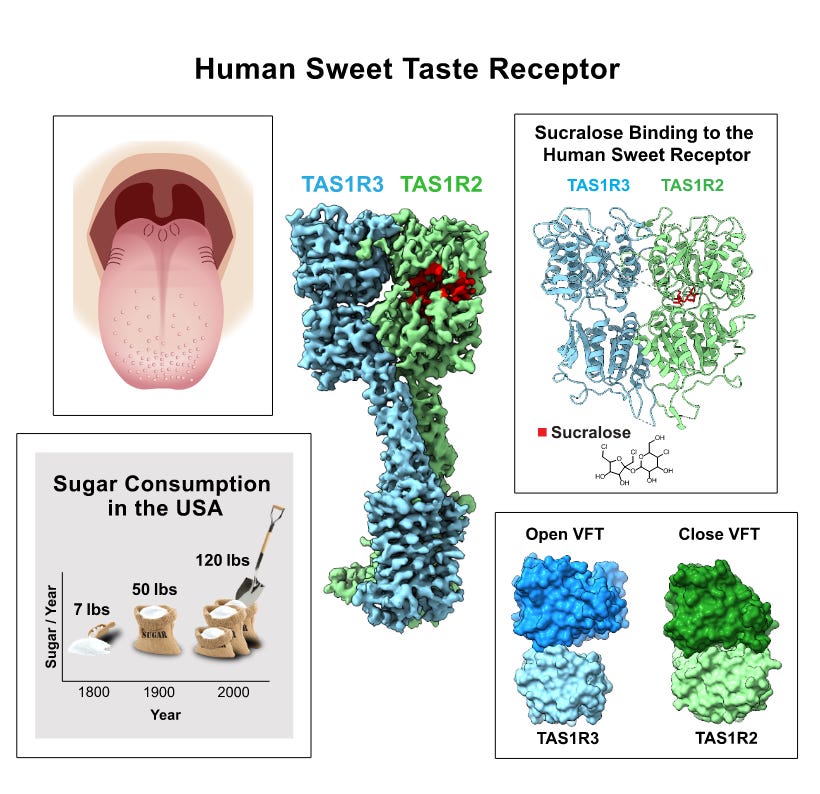
The structure of human sweetness [Juen et al., Cell, May 2025]
How does a single receptor mediate all of our responses to a wide range of sweet-tasting compounds?
The human sweet receptor is a heterodimer composed of TAS1R2 and TAS1R3 subunits. TAS1Rs are members of the GPCR family. The TAS1R2 subunit is unique to the sweet receptor, while TAS1R3 is also found in the mammalian umami receptor.
The human sweet receptor has two fascinating properties: it recognizes a broad, diverse set of natural agonists and is a low-affinity receptor (mM range for sugars). This makes intuitive sense as the role of the receptor is to endow animals with the capacity to recognize and differentiate between potential sugar sources; a low affinity receptor does not saturate at low concentrations, so confers the ability to choose richer sources of energy.
While the structure of many mammalian sensory receptors have been solved (e.g. temperature, mechanical, olfactory, bitter, itch, trace amine), the human sweet receptor has remained unsolved. Juen et al. from Columbia University used cryo-EM to determine the structure of the human sweet receptor bound to sucralose and aspartame. Aside from the important elucidation of the structure of the receptor, the authors found that the same residues on the TAS1R2 subunit appear to mediate the receptor-ligand interaction for both artificial sweeteners. Having the structure of the receptor might be able to help in the development of better sweet receptor modulators.
The time is ripe to reverse engineer an entire nervous system: simulating behavior from neural interactions [Haspel et al., arXiv, August 2023]
The Worminator paper begins with a striking comparison: just as chip designers painstakingly measure each transistor’s input–output curve to simulate an entire processor, neuroscientists could achieve true mechanistic insight by mapping every neuron’s state‐dependent IO‐function and replaying those functions in silico. If a “digital twin” of C. elegans can faithfully reproduce all its behaviors, from forward-backward crawling to feeding, egg laying and mating, then it has captured the true function of the nervous system.
The authors post the time is ripe to commence such a project, due to recent experimental breakthroughs. Automated microfluidic and robotic platforms can stage tens of thousands of age‐synchronized worms per year, while inverted SCAPE 2.0 light‐sheet microscopes sweep the entire 1 × 1 × 0.05 mm nervous system at up to 100 Hz, resolving all 302 somata simultaneously. Multicolor NeuroPAL strains, paired with AI‐driven alignment, tag each neuron across individuals, and stochastic optogenetic expression (where single‐cell activators light up random subsets) combined with whole‐brain calcium or voltage imaging, builds the complete nonlinear “interactome,” showing how any mix of inputs transforms into every neuron’s output.
One might wonder why it is really necessary to measure every single neuron’s activity. However, even neurons that look “redundant” at first glance can carry unique, state-dependent information and play non-obvious roles in network dynamics. For example, neuromodulators, intrinsic biophysics, and compartmentalized signaling can cause apparently redundant neurons to diverge under different conditions.
Beyond decoding the worm, this open‐science effort promises to drive advances in automation, data infrastructure, and neuromorphic AI by distilling the energy‐efficient, error‐robust computational primitives evolution has honed.
Notable deals
Azafaros has secured $147M in Series B funding in a round co-led by Jeito Capital and Forbion Growth.This financing will enable Azafaros’ lead product, nizubaglustat, to enter phase 3 studies for Niemann-Pick disease type C and GM1/GM2 gangliosidoses. Azafaros also plans to expand its pipeline into other indications.
Stylus Medicine has emerged from stealth with $85M from investors including RA Capital, Khosla Ventures, Chugai Venture Fund, Eli Lilly, J&J Innovation, the Myeloma Investment Fund and Tachyon Ventures. Stylus’ platform includes a therapeutic-grade recombinase, a therapeutic payload and a target LNP delivery system.
ABLi Therapeutics has emerged from stealth and is aiming to develop risvodetinib as a disease-modifying therapy for Parkinson’s and other diseases mediated by c-Abl.
Intrepid Labs has emerged from stealth and is aiming to transform drug formulation development through the use of AI and robotics. The company has raised a $7M seed round from AVANT Bio after already raising a $4M pre-seed round from Radical Ventures and Propagator Ventures. Intrepid Labs’ technology explores the full formulation design space to deliver optimized formulations.
Sphinx Bio has been acquired by Benchling. Sphinx has developed an AI-powered platform that helps life science labs automate complex and labour-intensive data work including parsing, analyses and transformations. Benchling and Sphinx aim to deliver a single, end-to-end platform for doing biology research where data and results are centralised to make decisions faster.
In case you missed it
Anthropic launches a program to support scientific research
CytomX Therapeutics Announces Pricing of $100 Million Underwritten Offering of Common Stock
What we liked on socials channels
Events
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @decodingbio.