BioByte 111: nanoparticles for brain drug delivery, AI tackles spatial biology, a new structure prediction model for designing protein binders, and the colossal undertaking of recreating the dire wolf
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
🚨 [Applications close 24th April] We're looking to expand the Decoding Bio team with Product Managers to transition Your Missing Guide to Breakthrough Biotech into a media empire. If you're interested in envisioning and implementing the future of Decoding Bio, fill out the quick form linked to the button below! 🚨
What we read
Blogs
The Process of Making a Dire Wolf [Colossal Biosciences, April 2025]
Just a little while ago we were intently watching as Colossal unveiled the incredible (and incredibly adorable) woolly mice, whose unique coats were the result of long-extinct woolly mammoth genes being spliced into the mouse genome. Now, the revolutionary company has realized something even bigger, the de-extinction of the dire wolf. In the past 6 months, Colossal has produced three healthy dire wolf pups, currently living on 2,000 acres of enclosed land, vigilantly monitored by a team of veterinarians.
Resurrecting an extinct species is no small feat, particularly given the fragility of DNA over the course of thousands of years. Many dire wolf skeletons exist thanks to the La Brea tar pits, but the heat of the tar penetrates the bones and destroys the DNA, making them largely unusable. A prior international collaboration examined 50 dire wolf samples, finding that only 2 had usable DNA. From these bones, they uncovered 0.1% of the dire wolf genome, something Colossal scientists significantly improved upon when they reexamined the samples.
With the knowledge of the relevant genes in hand, the scientists at Colossal took blood samples from grey wolves, an approach which significantly limits the detrimental impact to the wolves compared to more traditionally invasive techniques. Taking endothelial progenitor cells from the blood, which are more conducive to reprogramming, the scientists made edits across 14 genes to recreate the most relevant parts of the dire wolf genome. Using somatic cell nuclear transfer (SCNT)—the same technique used to clone Dolly the Sheep in the 90s—the nucleus of the edited cell was inserted into an egg cell with the nucleus removed. Surrogate mothers were then able to successfully birth a total of three dire wolves: Romulus, Remus, and Khaleesi.
Still, this development has naturally attracted skeptics. Many speculate whether these pups truly constitute dire wolves given the small number of genes that were incorporated and the fact that true dire wolves occupy an entirely different genus from today’s grey wolves. There are also concerns regarding the ethics of such experimentation and downstream effects should any of these new creatures be released into the wild. These fears echo the deleterious impacts of human-introduced invasive species and difficult quandaries with recent efforts around genetically engineering organisms such as mosquitos. Many of these points have been addressed by those at Colossal, who encourage the continuation of these open conversations and yet still emphasize the importance of their dogged pursuit of this science.
Ultimately, Colossal aspires to use the technologies they are developing to preserve the biodiversity of the planet as it continues to rapidly change, making edits that will help species survive in climates they did not originally adapt to. Their conservation work currently is focused on growing the population of the red wolf, only about 20 of which remain in the wild. Further advances in gene editing and splicing that Colossal develops will significantly aid them in this effort, and hopefully help prevent the projected 30% of genetic diversity loss expected by 2050. Furthermore, the noteworthy piece on Colossal’s breakthrough published by Time Magazine also highlights the decacorn’s two spin out biotechs, Breaking and Form Bio, which are focused on applying these groundbreaking techniques in genetic engineering to environmental pursuits of sustainable breakdown of plastic waste and drug discovery respectively. The myriad of applications are evident, and perhaps have never been more dire.
Papers
BoltzDesign I: Inverting All-Atom Structure Prediction Model for Generalized Biomolecular Binder Design [Cho et al., bioRxiv, April 2025]
Researchers at MIT and EPFL have developed BoltzDesign1—a model that repurposes Boltz-1 (an open-source reproduction of AlphaFold3) into a binder designer. This builds on previous work by the Correia and Ovchinnikov labs, who created BindCraft to transform AlphaFold2 into a protein binder designer with hit rates exceeding those of RFdiffusion. Unlike AlphaFold2, which is limited to proteins, both AlphaFold3 and Boltz-1 natively handle diverse biomolecules, enabling BoltzDesign1 to design binders for proteins, DNA/RNA, small molecules, and metal ions.
At a high level, BoltzDesign1 iteratively updates an input binder sequence so that Boltz-1 predicts strong binding to a target biomolecule. To avoid the high computational cost of backpropagating through the full diffusion module, the authors restrict optimization to the Pairformer. Instead of propagating gradients through the entire model, they backpropagate losses derived from the distogram and confidence metrics (pLDDT, pAE, pTM). Compared directly with RFdiffusionAA—a diffusion model that natively processes various biomolecules—this approach not only achieves superior performance in designing binders for four small molecules (IAI, FAD, SAM, OQO) but also produces protein binders with greater structural diversity.
Although explicit performance numbers aren’t provided, the authors indicate that BoltzDesign1 is more computationally efficient than RFdiffusionAA. Moreover, the model shows notable design flexibility: for instance, incorporating a loss function that penalizes alpha-helix formation can steer the design toward binders with more beta-sheet content. Overall, this work marks a significant advance toward the rational design of binders for a wide range of biomolecules, with promising implications for fields such as drug discovery, biosensing, and enzyme engineering.
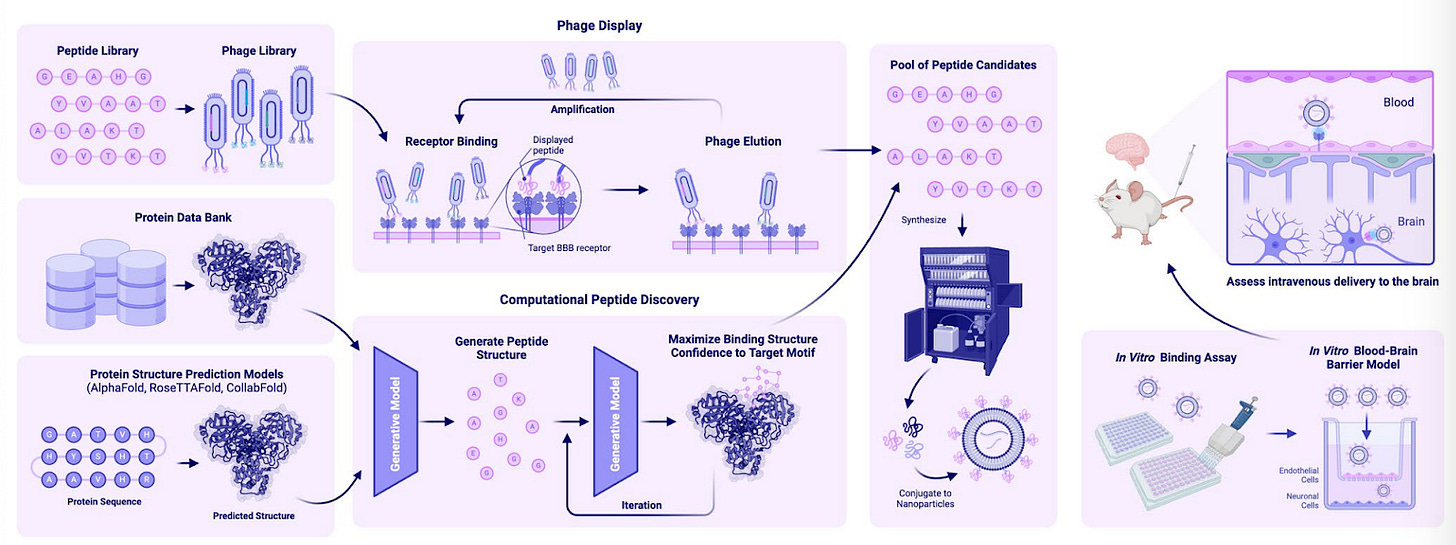
Peptide‐functionalized nanoparticles for brain‐targeted therapeutics [Tang et al., Drug Delivery and Translational Research, March 2025]
Nanoparticles (NPs) are useful vehicles for drug delivery allowing for prolonged circulation times, efficient transport through cell membranes, controlled release in target tissue and reduced side effects. Despite these advantages, delivery and biodistribution is limited due to clearance to the liver and spleen and presence of biological barriers. The blood-brain barrier (BBB) prevents 100% of all macromolecules from entering the brain parenchyma from the bloodstream.
NPs enter the brain via transcytosis across the brain endothelium. One way of increasing transcytosis is with targeting of receptors with high affinity ligands conjugated to NPs; facilitating receptor-mediated transcytosis (RMT). There are several issues with RMT including removal by efflux transporters, acidic degradation and unpredictable uptake routes. Antibodies, proteins, peptides and aptamers have all been used to bind to receptors on the BBB. Peptides, the review argues, are the optimal ligands for surface-functionalization due to their small molecular weights, easy synthesis and engineering, and low cytotoxicity and immunogenicity.
There are two main categories of peptides used to functionalize NPs:
Brain-targeting peptides: these include RVG29, T7, Angiopep-2 and mApoE which bind to receptors highly expressed on brain capillary endothelial cells (such as nAchR, GABARs, TfR, LRP1 and LDRLRs). They have demonstrated improvements in enabling transport across the BBB and cellular uptake leading to enhanced effectiveness for applications such as gene knockdown and gene editing.
Cell-penetrating peptides (CPPs): in order to be efficacious, many therapeutics need to enter the cell cytosol, so targeting the BBB is insufficient. To overcome this, NPs are functionalized with CPPs which are derived from proteins with transduction domains that can penetrate the cell membrane. TAT is a well studied CPP derived from the HIV-1 virus; when gold NPs are decorated with TAT, in vivo brain accumulation increases by 4.8-fold in GBM-bearing mice. Due to the untargeted nature of CPPs, they can also increase off-target effects unless they are co-functionalized with brain-targeting peptides.
As current brain-targeting peptides target highly expressed receptors in the brain that are also expressed in off-target organs, the authors suggest that computational peptide design has a great potential in generating de novo peptide binders to receptors specifically expressed in the brain or subpopulations of brain cells. These include receptors in the SREB family which have no known endogenous ligand and are known to modulate various neurological and psychiatric diseases.
SpatialAgent: An Autonomous AI Agent for Spatial Biology [Wang et al., bioRxiv, March 2025]
AI agents have the potential to accelerate complicated scientific workflows. However, to-date there have been few developments in automating complex workflows in spatial biology which requires specialized computational skills and substantial resources. In this preprint, the authors develop SpatialAgent, an LLM-based autonomous agent that can design gene panels for the experimental stage, conduct cell-type and tissue-niche annotation, summarize findings and generate hypothesis related to cell-cell interactions.
SpatialAgent integrates three core modules: memory, planning, and action. The action module handles technical tasks such as retrieving reference data, converting gene names, validating ligand-receptor interactions, processing data with existing tools, and generating or executing custom code. The memory module maintains both high-level objectives (semantic memory) and task-specific progress (episodic memory). The planning module employs chain-of-thought reasoning and self-reflection to break down complex goals into executable steps.
First, SpatialAgent was tested in designing gene panels. SpatialAgent outperformed established computational pipelines and enhanced human-designed gene panels. The authors also show that SpatialAgent can effectively automate and standardize cell and tissue niche annotation. SpatialAgent could also extract biological insights including identifying key signaling pathways and distinct spatial transcriptomic signatures. Finally SpatialAgent was used to support the design of an ongoing experiment enhancing the discovery of cell-type specific communication patterns.
This work shows how autonomous AI agents can save time and accelerate the discovery of novel biology.
Synthesizing world models for bilevel planning [Ahmed et al., arXiv, March 2025]
In the realm of reinforcement learning (RL), video games have long served as a proving ground for novel approaches that seek to decode human-like intelligence. Modern deep RL agents have achieved impressive feats but remain starkly inefficient compared to the way humans quickly grasp new games or environments. Traditional agents often learn through massive amounts of trial and error without the benefit of a deeper, causal understanding of their world. TheoryCoder enters this scene by mimicking the way humans build internal models of their surroundings, constructing structured, high-level abstractions and grounding them through program synthesis. This approach leverages the conceptual clarity of planning languages and the expressive power of Python to simulate and refine world dynamics from minimal experience.
This development matters because it challenges the traditional view of learning solely through direct experience. By integrating high-level planning with low-level action synthesis, TheoryCoder paves the way for more efficient exploration and faster adaptation in complex, dynamic domains. Its success in grid-world puzzles illustrates how drawing on human-inspired structures can lead to higher sample efficiency, improved generalization, and a deeper understanding of the environment. (overview of TheoryCoder below.)
Notable deals
Merida Biosciences, founded by Third Rock Ventures in 2022 and based in Boston, raised $121 million in Series A funding led by Bain Capital and BVF. The company will start clinical trials this year for an experimental Graves’ disease therapy, with potential use in thyroid eye disease, and plans to advance treatments for food allergy and primary membranous nephropathy. Its broader goal is to neutralize pathogenic antibodies underlying multiple diseases.
Exobiosphere has secured a €2M seed round to build the world’s first contract research organization dedicated to drug discovery in space, with Expansion Ventures leading the investment. The funding will support the development of the Luxembourg-based company’s Orbital High Throughput Screener, a platform specifically designed to operate in microgravity. According to the company, microgravity induces cellular effects that cannot be replicated on Earth, such as accelerated cellular senescence, neurodegeneration, and faster neuronal development. These unique properties offer promising opportunities for more efficient disease modeling and a deeper understanding of drug therapeutics within shortened testing timelines.
Neurona Therapeutics has raised $102M to fuel its Phase III epilepsy trial and advance its suite of regenerative neural cell therapies. With participation from Fidelity, Viking Global, and Cormorant, the funding will accelerate the development of off-the-shelf, allogeneic neural cell therapy candidates engineered for single-dose, long-term repair of the nervous system. The company's lead product candidate, NRTX-1001, is composed of GABAergic interneurons derived from a unique medial ganglionic eminence lineage that plays a critical role in brain development by generating inhibitory neurons and myelinating glial cells essential for proper neural circuit function. This innovative approach aims to provide targeted repair of neural circuits affected by chronic neurological disorders, addressing the shortcomings of traditional therapies that often exhibit imprecise and diffuse effects.
RayThera raises $110M Series A to advance its suite of preclinical immunology candidates through Phase I. The round was led by Foresite Capital and Orbimed.
Atsena Therapeutics, a clinical-stage gene therapy company focused on reversing or preventing blindness, has closed a $150 million Series C round led by Bain Capital’s Life Sciences team with participation from Wellington Management and existing investors. The funding will accelerate the development of ATSN-201 for treating X-linked retinoschisis and bolster the company’s preclinical pipeline of innovative ocular gene therapies.
What we listened to
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here.

















The discussion on nanoparticles for brain drug delivery is particularly intriguing. Overcoming the blood-brain barrier has long been a significant challenge in neuropharmacology. The advancements highlighted here suggest we’re making meaningful progress. Additionally, the integration of AI in spatial biology and the development of new protein structure prediction models underscore the interdisciplinary nature of current biotech innovations. It’s exciting to see how these converging technologies are paving the way for more effective therapies.