BioByte 110: a blood test for diagnosing Alzheimer's, future behavior encoding in rats, entropy-reinforced planning in LLMs for drug discovery, & an exciting call to join Decoding Bio!
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
🚨 We're looking to expand the Decoding Bio team with Editors and Product Managers to transition Your Missing Guide to Breakthrough Biotech into a media empire. If you're interested in envisioning and implementing the future of Decoding Bio, fill out the quick form linked to the button below! 🚨
What we read
Blogs
New Alzheimer’s blood test accurately tracks disease progression [Eleanor Garth, Longevity.Technology, April 2025]
A collaborative study between researchers at Washington University School of Medicine in St. Louis and Lund University in Sweden has shown the powerful capacity for tracking Alzheimer’s disease by measuring specific plasma biomarkers in a simple blood test. Previously only diagnosable through costly PET scans, screening for the presence of MTBR-tau243 in the blood is en route to enabling greater diagnostic capabilities at a fraction of the time and cost. MTBR-tau243 is a tau protein fragment existing in the bloodstream; its value as a biomarker of Alzheimer’s derives from its strong propensity to reflect the accumulation of tau protein tangles in the brain, a hallmark of the indication. This measure of elevated tau is distinct from detection of amyloid plaques, which, unlike tau tangles, can precede cognitive symptom expression. The elevated levels of MBTR-tau243 in the bloodstream were also shown to be highly correlated with symptomatic disease progression, even demonstrating stronger linear correlations than PET imaging in later stages of the condition.
Both of these traits of the MBTR-tau243 biomarker confer enormous advantage for clinicians developing treatment plans, especially when utilized in tandem with tests for plasma levels of p-tau217, a biomarker with a greater aptitude for ascertaining earlier stages of the disease (i.e. before symptomatic presentation) via detection of amyloid plaques. These tests utilized together exhibit high confidence in elucidating Alzheimer’s as the underlying pathology in patients and inform treatment as well as significantly augment and better select participants for clinical trials. For example, the article highlights the effectiveness of anti-amyloid therapies such as lecanemab in early stages of the disease when tau burden is low, as well as the subsequent shift to tau-targeting therapies at later stages. Such understanding can lead to much improved efficacy of treatment and better patient outcomes.
This breakthrough may very well herald the new era of ‘personalized medicine for Alzheimer’s’ signifying both a substantial scientific as well as social shift, especially as further comprehension of the nuances of this widespread and debilitating indication continue to unfold. While this discovery still requires further validation and, at present, necessitates specialty mass spectrometry equipment and sizable plasma volumes for testing, its development as a widely accessible blood test in the near future is nevertheless quite feasible—a critical mark of progress toward sustained health as longevity increases.
Papers
Constructing future behavior in the hippocampal formation through composition and replay [Behrens et al., Nature Neuroscience, March 2025]
A key question in computational neuroscience is how the hippocampus, known for its role in memory and spatial navigation, can quickly build flexible maps of the world without having to learn every detail from scratch. In this paper, researchers from Oxford and other institutions propose that the hippocampus constructs these maps by composing them from reusable building blocks that are already represented in the cortex. These building blocks include grid cells, which signal where you are, and object vector cells, which indicate where other important features are relative to you.
The model suggests that the hippocampus binds these building blocks together into conjunctive memories that represent space and guide behavior. Replay, a process in which the brain simulates sequences of neural activity while at rest, plays a critical role by consolidating these compositional memories. The authors provide computational simulations and analyze hippocampal recordings in rats to show that replay events are associated with the formation of new place fields. For example, when a rat replays a location where a new reward or a structural change such as a locked door is present, a new neuronal response can emerge that guides future behavior.
This work matters because it offers a unifying explanation for how the brain can support rapid zero shot generalization to new environments. By composing maps from familiar elements, the hippocampus can efficiently update its representation of the world and guide behavior immediately, even without direct experience in a novel situation. This insight not only deepens our understanding of memory and navigation but may also influence the design of artificial intelligence systems that need to generalize learning across different contexts.
A light-inducible RNA base editor for precise gene expression [Li et al., Nature Biotechnology, March 2025]
A major challenge in gene therapy is the precise control of when and where therapeutic edits occur, which is essential for improving both safety and efficacy. To tackle this, researchers have developed a photoactivatable RNA adenosine base editor (PA-rABE).
The PA-rABE system consists of a split ADAR2 enzyme fused to either a positive or negative Magnet domain. Upon exposure to blue light, the Magnet domains dimerize leading to two halves of the ADAR2 protein reassemble to form a functional enzyme. In this way editing only occurs when the editor is exposed to the correct light source.
To optimize the system, the researchers tested various constructs of the ADAR2 and Magnet domains. They then evaluated PA-rABE’s ability to correct nine different disease-causing mutations, achieving editing efficiencies ranging from 12% to 42%. When compared to other RNA editors, PA-rABE demonstrated superior editing rates and reduced off-target activity.
PA-rABE was also tested in vivo. First, the authors tested the ability of the PA-rABE to restore the activity of firefly luciferase in mice. Mice that were exposed to blue light had a dramatically elevated level of luciferase reporter activity compared to controls. Second, PA-rABE was tested in hemophilia B mice showing effective restoration of target gene expression and an improvement in coagulation function.
These results suggest that PA-rABE provides a new way to achieve effective spatial and temporal control over RNA editing.
Entropy-Reinforced Planning with Large Language Models for Drug Discovery [Liu et al., arXiv, March 2025]
The authors of the paper describe a new algorithm named Entropy-Reinforced Planning (ERP), designed to improve transformer models in their use of molecule generation for drug discovery. ERP addresses limitations of LLM-based molecule generation methods by better balancing exploration versus exploitation during token selection.
ERP integrates Monte Carlo Tree Search (MCTS) with entropy values to guide the transformer’s decoding process, leading to higher quality and more diverse molecules. The entropy term evaluates the uncertainty in the probability distribution of tokens predicted by the transformer at each step; which helps identify areas of high uncertainty in molecular space, rewarding exploration of less-explored regions that may contain high-reward molecules.
ERP demonstrated that it outperformed across four baselines (Beam Search, Sampling, UCT, PG-TD) for several properties (Docking, Druglikeliness, Synthesizability, Solubility) evaluated on the SARS-CoV-2 virus (3CLPro) and human cancer cell target protein (RTCB) benchmarks:
Benchmarking and optimizing organism wide single-cell RNA alignment methods [Díaz-Mejía et al., arXiv, March 2025]
Single-cell RNA-seq studies are piling up, but aligning them into a unified map of biology is still messy. Most models are evaluated on small datasets with metrics that don’t capture the tradeoff between correcting batch effects and preserving biological signal. This paper takes a step toward solving that with two key contributions: a new benchmark (think MNIST and ImageNet, but for scRNA data) and a tougher, more interpretable metric called the K-Nearest Intersection (KNI) score.
The KNI score works at the datapoint level, combining batch mixing (via kBET) with accuracy at predicting cell-type labels—rewarding models that do both well, not just one. Using this setup, the authors introduce BA-scVI, a 4-layer adversarial variant of scVI that consistently outperforms existing methods across both small (scMARK) and large (scREF) benchmarks.
Some notable findings:
Transformer models like scGPT and geneFormer don’t perform well out of the box. They tend to lock in batch effects during pretraining, making fine-tuning for alignment harder unless batch effects are explicitly handled during training.
BA-scVI works just as well without injecting batch IDs into the encoder, making it well-suited for inference and reference-based alignment of new data.
Supervised models are dangerously good at overfitting. Even slight exposure to cell-type labels leads to misleadingly high performance. The authors argue we should stick with unsupervised approaches that only see technical variables (e.g., batch/study ID) during training.
The result is a scalable, high-performing unsupervised method that preserves fine-grained biological detail while aligning data across technologies and tissues — a key capability for building organism-wide cell atlases. And the KNI metric gives us a much-needed way to tell when models are actually doing the right thing.
Notable deals
Siemens AG is set to acquire Dotmatics, a leading provider of life sciences R&D software, for $5.1 billion. This move extends Siemens’ AI-powered ‘Product Lifecycle Management’ portfolio into Life Sciences, aiming to seamlessly connect research and development with manufacturing. The acquisition expands Siemens’ industrial addressable software market by $11 billion, reinforcing its strategy to accelerate customer innovation across industries with high R&D spend.
AIRNA raises $155 million in Series B funding to advance its alpha one antitrypsin deficiency program into the clinic later this year. The candidate AIR 001 repairs faulty RNA produced by the mutated gene in Alpha-1 antitrypsin deficiency, known as AATD. When the RNA is edited, cells generate functional copies of the AAT protein, reducing the risk of lung and liver disease associated with this condition. The Series B round was co-led by Venrock and Forbion.
Galatea Bio has raised $25 million to build a global biobank of 10 million people, targeting the discovery of disease drivers, new drug targets, and biomarkers. The initiative emphasizes including underrepresented populations to overcome the current bias toward data from individuals of European descent. Inspired by deCODE Genetics' success, which mined genetic data to identify disease risks and led to a $415 million acquisition by Amgen, Galatea Bio will leverage a machine learning first approach and forge global partnerships, including in Africa. The diverse dataset could uncover novel genetic variants that lead to treatments benefiting all populations, as exemplified by rare cases like the Nav1.8 mutation causing congenital insensitivity to pain.
Isomorphic Labs, an artificial intelligence powered drug discovery company spun out of DeepMind, has raised $600 million in its first external funding round led by Thrive Capital with participation from GV and Alphabet. The fresh capital will propel the company to invest in small molecule drugs and antibodies while exploring a US office launch as it prepares to enter clinical trials. The firm is focused on enhancing data curation and generation to accelerate its advancing drug design programs and deliver innovative therapies to patients.
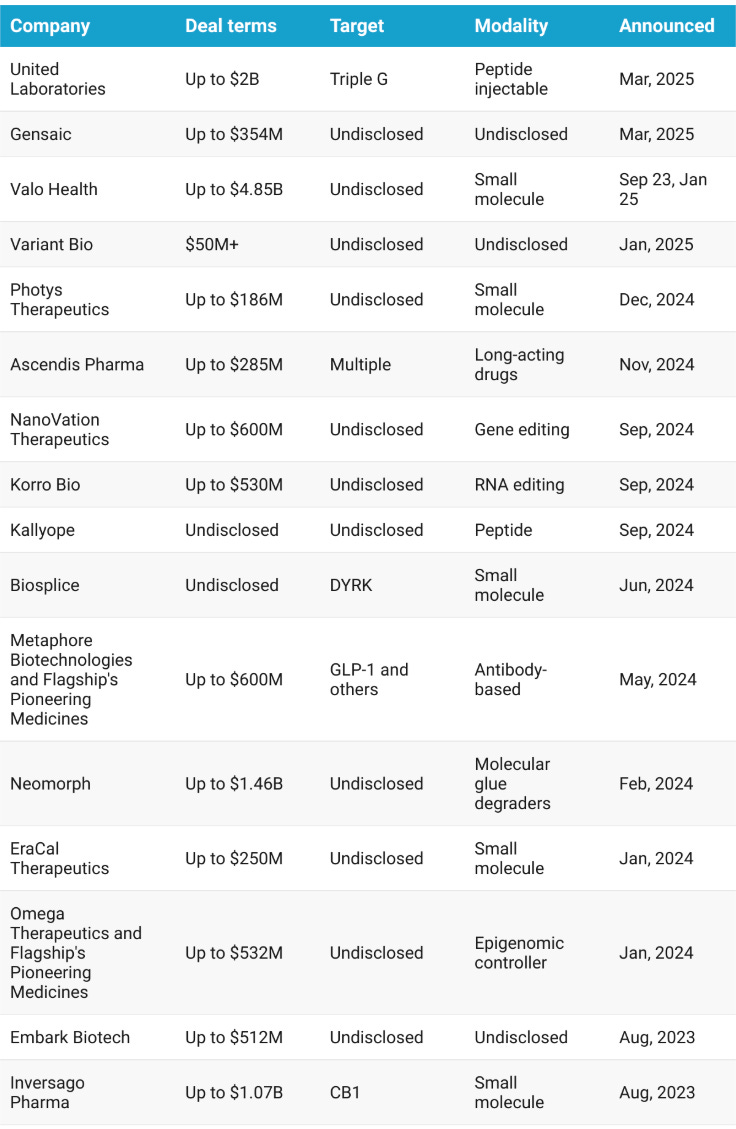
Novo signs its second obesity deal in a week, picking up Lexicon’s oral small molecule LX9851 for $75 million upfront and up to $1 billion in milestones. The drug targets ACSL5, a metabolic enzyme tied to fat build-up and energy regulation, and is still preclinical. The idea is to help people feel full and slow gastric emptying.
Lexicon’s asset is first in class with a completely novel mechanism. Novo gets global rights and a shot at a differentiated oral therapy that could either complement existing GLP-1s or stand alone as part of what Lexicon’s CEO calls the obesity 2.0 treatment paradigm. It is a strong price for Novo, especially given Lexicon’s position. The company’s stock is down about 80 percent over six months, with recent trial misses, layoffs, and ongoing struggles to get its heart failure drug approved by the FDA. Novo, meanwhile, is racing to build its next obesity franchise before semaglutide loses exclusivity in the early 2030s. This deal adds to a growing list of bets on new mechanisms and long-term cardiometabolic presence.
Above: Select novo obesity deals (source: Endpoints)
What we listened to
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here.















I appreciate these Bytes! My humble vote for the next Field Trip :) https://www.youtube.com/watch?v=wneX5-M07TA