BioByte 105: sequencing by expansion, self-driving labs for novel lipid discovery, mass spec proteomics, and a return to Asilomar
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
Mass-spectrometry-based proteomics: from single-cell to clinical applications [Guo et al., Nature Reviews, February 2025]
A new Nature review highlights the latest developments in mass-spectrometry (MS)-based proteomics, a field that is now capable of delivering unprecedented insights at both the single-cell level and within complex tissue environments.
Recent technological breakthroughs—including higher sensitivity instrumentation, improved throughput, and AI-driven data analysis—have transformed proteomics from a niche research tool into a powerful approach for mapping cellular heterogeneity, characterizing post-translational modifications, and identifying protein–protein interactions. Beyond fundamental research, MS-based proteomics is making significant strides in clinical applications. Advances in biomarker discovery from body fluids, such as plasma and urine, are paving the way for earlier disease detection and more precise diagnostics. The development of targeted proteomics assays is further accelerating the transition of proteomics into clinical practice, offering new possibilities for personalized medicine that solely looking at genetics doesn’t provide.
AI-powered tools are a big factor in the advance of proteomics—helping with refining protein identification, quantification, and functional interpretation of complex datasets. As these models continue to evolve, they hold the potential to streamline biomarker discovery, optimize therapeutic development, and integrate proteomics with other multi-omics approaches for a more comprehensive understanding of biology.
Decoding the proteome will allow for the full potential of personalized medicine, pushing on the advances we’ve seen in the last two decades from genomics.
50 years after a seminal conference, big questions about biotechnology remain [Nell Greenfieldboyce, NPR, February 2025]
Fifty years ago this week, a group of 150—comprised primarily of scientists—gathered at Asilomar State Beach to discuss the new directions and applications of recombinant DNA technology, as well as the risks entailed with this breaking of new ground within genetic experimentation. Those in attendance at the historic Asilomar Conference were focused on the outcomes that could result from splicing DNA of different species, specifically what the various applications of this technology could mean for the discipline and the world at large. The handful of lawyers in attendance emphasized the gravity of the scientists themselves cultivating standardized, safe practices to guide this burgeoning field of research, as others who would step in if they abstained would not be as informed or enthused about the subject. This original conference is regarded as a marked success, as much of what was conceptualized at this meeting in 1975 laid the groundwork for the legislation still enduring today.
In celebration of this pivotal event, another conference was held earlier this week, echoing sentiments around pioneering science from the original conference, although now applied to even further-reaching frontiers. Much has changed as subsequent technological developments have significantly progressed in the field. With it now flourishing in full-swing, there are many more working in this area than ever before, pursuing an even greater multitude of directions. It is both an extremely exciting time as well as perhaps one of the most dangerous to date, with possible implications embodying much more extensive and dire consequences to the planet and its inhabitants. The gravity of these considerations render it even more pertinent to revisit old questions about ethics and risks as well as broach new ones. While scientists at the original conference largely concentrated on recombinant DNA, those in attendance at the most recent are engaged with even more radical initiatives such as the idea of mirror life.
Differing from the conference in 1975, however, this year there was a special initiative to include a greater number of younger scientists within the event, highlighting the importance of including newer voices and fresh outlooks in these conversations. With this arsenal of brilliant minds, diverse perspectives, as well as the continuing incredible advancements in biotech and the ensuing proactive debate around their ethical implications, there are certain to be even more exciting things to come in the next 50!
Papers
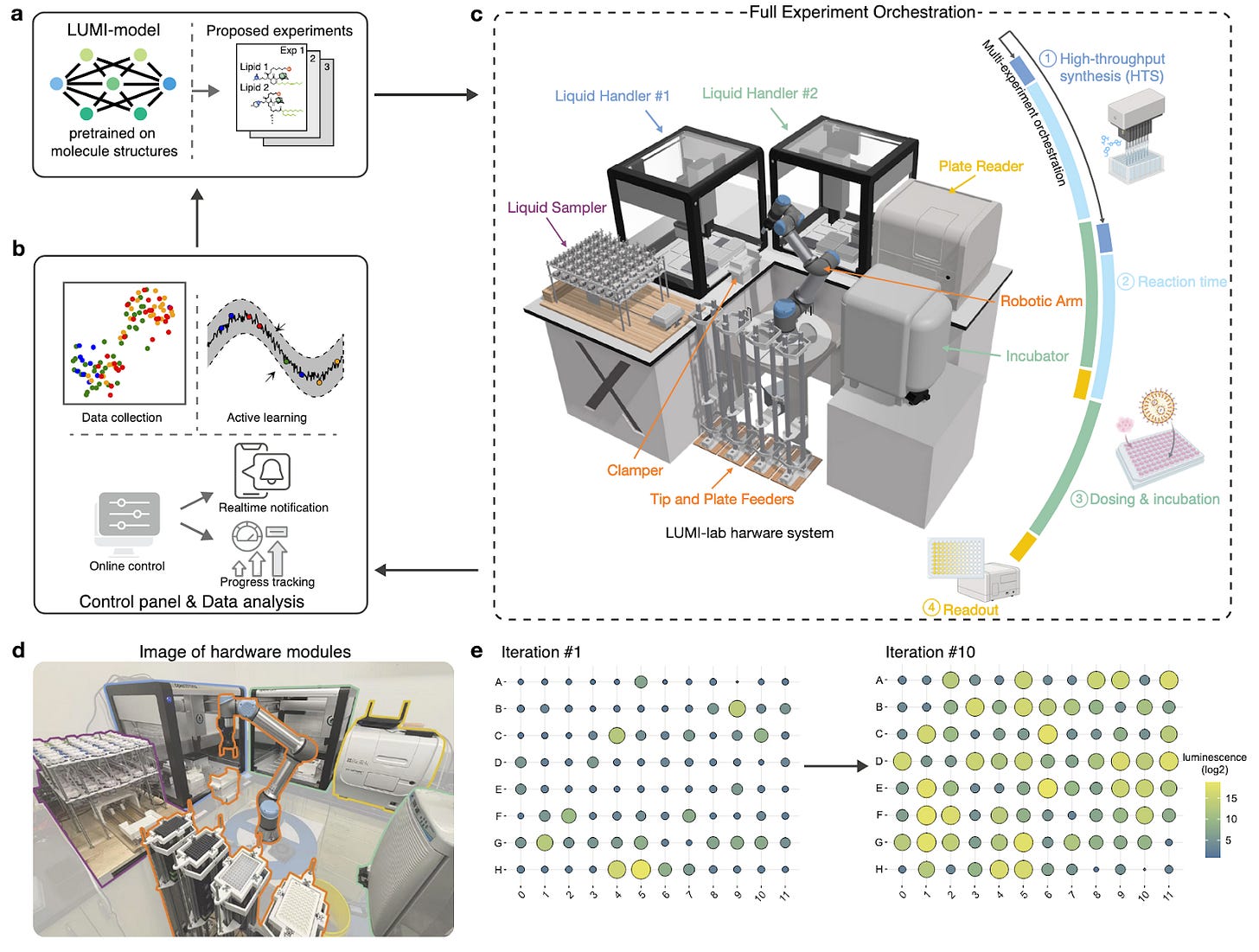
LUMI-lab: a Foundation Model-Driven Autonomous Platform Enabling Discovery of New Ionizable Lipid Designs for mRNA Delivery [Cui et al., bioRxiv, February 2025]
The development of self-driving labs (SDLs) remains a particular challenge in fields with a scarcity of annotated historical data, which makes it difficult for models to learn and adapt to unexplored chemical spaces.
The issue of data scarcity is clear in the development of new lipid nanoparticles (LNPs), such as the ones used in mRNA vaccines. Data is scarce as there are only three LNPs that have received FDA approval to date. The design of one of the critical components of the LNP, the ionizable lipid, which is crucial for encapsulation and endosomal escape, has relied on expert knowledge due to the lack of systematic development platforms.
The authors set out to build an SDL to identify new ionizable lipids named LUMI-lab (Large-scale Unsupervised Modeling followed by Iterative experiments). It consists of two primary components: 1) LUMI-model, a molecular foundation model pretrained with unsupervised learning on 28 million structures, which facilitates a few-shot learning of new molecules with minimal experimental data and 2) an automated, closed-loop experimental system that generates wet-lab data to optimize LNPs through ionizable lipid synthesis (via OpenTrons OT-2), LNP formulation, and high-throughput in vitro screening.
In ten iterative cycles, LUMI-lab synthesized and evaluated >1700 new LNPs, with new ionizable lipids with superior mRNA transfection potency (mTP) in human bronchial epithelial cells. Five out of the top identified lipids demonstrated mTP comparable or exceeding SM-102 (the LNP used in Moderna’s COVID-19 vaccine). The team also demonstrated that one of the new lipids increased epithelial cell editing efficacy in the lung via LNP-mediated delivery of CRISPR-Cas9 via inhalation; surpassing the highest editing efficiency previously reported.
An opponent striatal circuit for distributional reinforcement learning [Lowet et al., Nature, February 2025]
A new study in Nature explores how the brain encodes not just average rewards but the full range of possible outcomes, revealing a more nuanced approach to decision making. Researchers at Harvard identified distinct groups of neurons in the striatum that separately encode better than expected and worse than expected rewards. This aligns with distributional reinforcement learning (RL), a machine learning approach that improves decision making by considering full reward distributions rather than simple averages. Using high density neural recordings in mice, the study found that dopamine inputs help organize these distributional representations, suggesting that our brains naturally compute a richer picture of risk and reward. These findings could enhance our understanding of human decision making, particularly in conditions where reward processing is disrupted, such as depression or addiction.
Sequencing by Expansion (SBX) -- a novel, high-throughput single-molecule sequencing technology [Kokoris et al., bioRxiv, February 2025]
Roche has unveiled Sequencing by Expansion (SBX), a novel single-molecule sequencing technology. SBX has been developed to address the fundamental signal-to-noise limitations of nanopore sequencing that arise from the difficulty of distinguishing closely spaced nucleotide bases. SBX consists of two separate steps: synthesis and sequencing.
Synthesis. The sequence of a target nucleic acid molecule is encoded into an expanded surrogate polymer called an Xpandomer. This converts the DNA into a more measurable structure for the nanopore. An Xpandomer consists of expandable nucleoside triphosphate substrates (XNTPs) which are added sequentially by a highly engineered DNA polymerase (the Xp synthase). Each XNTP encodes the base identity of the cognate nucleotide with a modified dNTP and also consists of a reporter, translocation control element (TCE) and two enhancers. The dNTP is attached to each enhancer in two separate places such that the reporter loops round creating a circularised molecule. After all the XNTPs have been added the sugar-phosphate links are degraded with acid. This breaks open the circular XNTPs resulting in an opening up of the Xpandomer into a single linear molecule around 50-times the length of the target DNA.
Sequencing. The Xpandomer is sequenced by stepwise translocation through a nanopore. Here, the translocation control element plays a crucial role, holding the reporter sequence in the nanopore while the ion current is measured. Each base has a corresponding reporter that yields a distinct, differentiated ion current level. A high-voltage release pulse results in translocation of the translocation control element through the nanopore and measuring of the subsequent reporter.
A cool animation is included in the press release here. This is an awesome feat of engineering involving the generation of novel enzymes, molecules and synthesis routes.
Notable deals
Eikon Therapeutics has secured a $351 million Series D financing, bringing its total funding to over $1.1 billion since 2019. This round was raised at a lower valuation than its $3.6 billion Series C, reflecting broader market trends even for well-progressing biotechs. The investment will support the company’s drug discovery efforts, which leverage single-molecule tracking technology to analyze proteins in living cells with extraordinary precision. Eikon’s lead candidate, EIK1001, a toll-like receptor co-agonist, has entered Phase 3 evaluation in advanced melanoma, while EIK1003, a selective PARP1 inhibitor for breast, ovarian, prostate, and pancreatic cancers, is in Phase 1. Both programs were in-licensed to jumpstart the company’s pipeline, which also includes a brain-penetrant PARP1 inhibitor (EIK1004) and an internally derived WRN inhibitor (EIK1005).
Achira launches with $33 million in funding from NVIDIA’s venture arm, Amplify, Compound, and Dimension to bring neural network potentials (NNPs) to drug discovery. NNPs are machine learning models that approximate the Schrodinger equation, combining the accuracy of quantum mechanics with the speed of classical molecular mechanics. By enabling more accurate binding affinity predictions, NNPs can help streamline the development and optimization of promising drug candidates, a critical step in pharmaceutical research. The company was co-founded by Zavain Dar (Dimension), John Chodera, and Theofanis Karaletsos (CZ Biohub). While Achira is entering an emerging and promising space, there is a lot of technical risk to address and its business model has not yet been specified.
Sortera Bio, a spin-out from the LMB, has officially emerged from stealth to accelerate generative AI-driven drug discovery with its ultra-high-throughput Deep Screening platform. The company’s Deep Screening technology enables the rapid generation of massive, unbiased experimental datasets. Deep Screening integrates massively parallel sequencing, ribosome display, and affinity display to generate hundreds of millions of sequence-function pairs per experiment. This enables the rapid identification of high-affinity antibodies against clinically relevant targets. By combining Illumina’s next-generation sequencing with ribosome display, the method allows for the simultaneous testing of millions of antibody-antigen interactions within a single experiment.
Enveda Gains Backing from Sanofi to Advance AI-Driven Drug Discovery to Clinical Trials Bringing Total Series C Financing to $150M. Following its Series C announcement, Sanofi has entered the syndicate to back the company's I&I pipeline, which includes a first-in-class molecule for atopic dermatitis and asthma in the clinic, and its metabolite-driven drug discovery capabilities.
Vevo Therapeutics has open-sourced its 100M cell dataset contributing to the Arc Institute’s Virtual Cell Atlas. This dataset captures 60,000 drug-patient interactions from perturbation experiments, mapping the responses of 50 diverse cancer models to 1,100+ drug treatments.
What we liked on socials channels
What we listened to
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here.