BioByte 104: the release of Evo 2, validating free-form hypotheses at scale, shedding light on astrocytes, and a guide to biotech partnerships
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
The Practical Guide to Biotech Partnerships [Lucas Harrington, Feb 2025]
In a comprehensive guide drawing from his experience at Mammoth Biosciences, Lucas Harrington shares hard-earned insights on navigating biotech partnerships from initial courtship to long-term collaboration. His perspective is particularly relevant as challenging equity markets make strategic partnerships an increasingly critical lifeline for biotechs.
The heart of Lucas’ message is that successful partnerships are more like marriages than transactions—building relationships that can weather inevitable challenges and capitalize on unexpected opportunities is key.
Some insights:
Partnership timing requires careful strategy: while asset value typically increases with development progress, companies must find the sweet spot between investment and partnership value. These decisions have years-long implications.
Leverage your network aggressively for senior-level introductions
Use conferences and publications strategically for inbound interest
Track all interactions meticulously—partnership conversions often take years
Build relationships early, even before you're ready to partner
Success hinges on a passionate internal champion at the partner organization and competitive tension in the process
Lucas warns against common pitfalls like letting legal teams dominate business discussions or manufacturing artificial urgency with partners. He emphasizes that while contracts matter, relationship and incentive alignment matter more: "A good partner with a bad contract can still succeed, but a bad partner with a good contract is destined for failure."
For platform companies, the key takeaway is selectivity—choosing partnerships where both parties share a genuine commitment to long-term collaboration. The most valuable partnerships aren't necessarily those with the highest upfront payments, but rather those where strategic alignment creates opportunities for both companies to grow and innovate together.
Papers
Uniform volumetric single-cell processing for organ-scale molecular phenotyping [Nature Biotechnology 2025]
Mouse brain hemisphere is stained with various cell type markers: neurons overall (cyan), and cells specifically involved with neurotransmitters dopamine (yellow) and acetylcholine (magenta)
Single-cell analysis has dramatically advanced our understanding of cellular diversity and function, but applying these techniques to intact, organ-wide tissues remains a formidable challenge. A major bottleneck in single-cell studies is achieving uniform chemical processing across densely packed cells within large tissues. Conventional methods struggle to ensure that all cells, from surface layers to deep tissue regions, receive equal exposure to chemical reagents, leading to inconsistencies in labeling and data quality.
Historically, two main approaches have been used to tackle this issue:
Tissue Dissociation – Breaking tissues down into individual cells allows for even chemical exposure but destroys spatial context, making it impossible to analyze cell-to-cell interactions within an intact organ.
Thin Sectioning – Cutting tissues into ultra-thin slices improves accessibility for chemical reagents, but loses the full 3D architecture of the organ, limiting the ability to study large-scale structural patterns.
Attempts to label whole organs while maintaining spatial integrity have relied on diffusion-based techniques. However, these methods suffer from uneven penetration of reagents, especially in thick tissues, resulting in incomplete labeling and biased results.
What's new?
A new study introduces CuRVE, a framework designed to actively maintain a uniform chemical environment within intact tissues. Unlike traditional methods that rely on diffusion, CuRVE continuously modulates reaction conditions, ensuring that all cells experience equal exposure to reagents, regardless of their location within the organ.
To implement CuRVE practically and at scale, the researchers developed eFLASH (electrophoretic-Fast Labeling using Affinity Sweeping in Hydrogel), which combines electrophoresis with controlled pH and detergent sweeps to enhance reagent penetration.
This has so far enabled:
Whole mouse and rat brains immunolabeled within one day - a process which can usually take weeks.
Uniform single-cell labeling across entire organs, eliminating the variability seen in traditional diffusion-based approaches.
Discovery of previously undetected biological phenomena, such as region-specific reductions in parvalbumin-positive (PV+) cells in wild-type mouse brains, a phenotype missed by genetic labeling techniques.
Astrocyte ensembles manipulated with AstroLight tune cue-motivated behavior [Serra et al., Nature Neuroscience, February 2025]
Neurons receive the majority of attention in studies of brain function and organization. However, these studies are not always able to capture the nuances of individual neuronal circuits, largely because they are not able to identify and manipulate the role and function of non-neuronal cells, astrocytes in particular. Serra et al. present and validate a method to study and stimulate astrocytes using a technique they dub AstroLight, an adaptation of the Cal-Light dual-protein switch system.
AstroLight uses three viral vectors to identify and subsequently control astrocytes that are activated during a specific behavioral task. When exposed to blue light and a higher concentration of calcium ions, two vectors work together to release a promoter that is able to induce transcription of a fluorescent-tagged protein of interest. This inducible expression enables the authors to monitor and manipulate activity of these task-associated astrocytes, reactivating them or introducing gain-of-function and loss-of-function alterations. In this study, the authors elected to use AstroLight to investigate the role of astrocytes in the nucleus accumbens circuit (NAc)—an area of the brain associated with reward-seeking, decision-making, and spatial memory—due to the dearth of investigations of non-neuronal components in this area.
Upon building a cue-reward relationship in AstroLight-engineered mice, the authors discovered an increased recruitment of astrocytes in the NAc. They also found this set of task-associated astrocytes to strongly influence choice. When mice were presented with a set of identical cues, those with astrocyte activity induced by AstroLight preferentially engaged with one. Astrocyte gain-of-function and loss-of-function alterations replicated these results, with the gain-of-function mice demonstrating asymmetric interest and the loss-of-function displaying indifference.
Through these results, Serra et al. demonstrate that astrocytes in the NAc are organized into distinct functional clusters, with some encoding information specific to a discrete stimulus, behavior, and reward. These results highlight the potential of Astrolight to unlock the intricacies of neural circuits, shining a light on previously overlooked components.
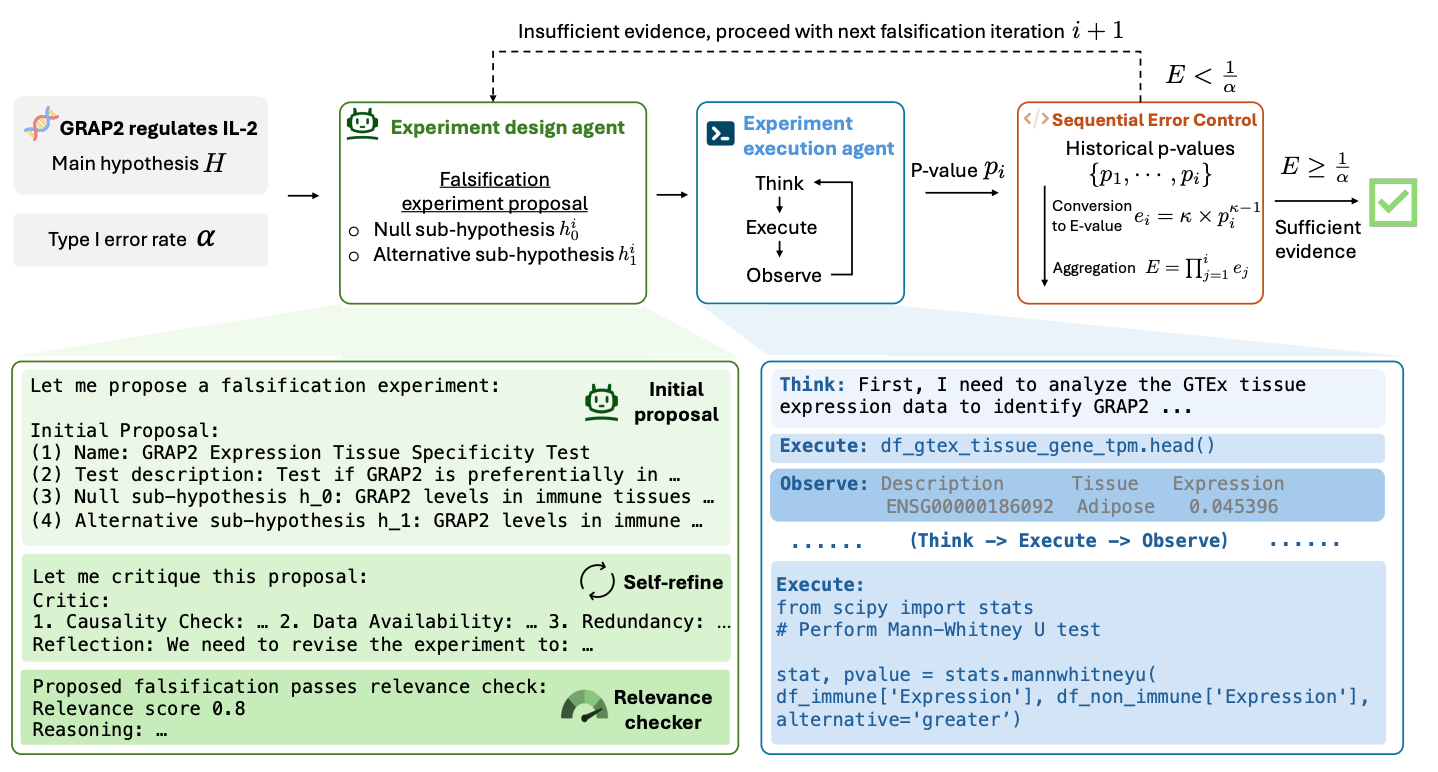
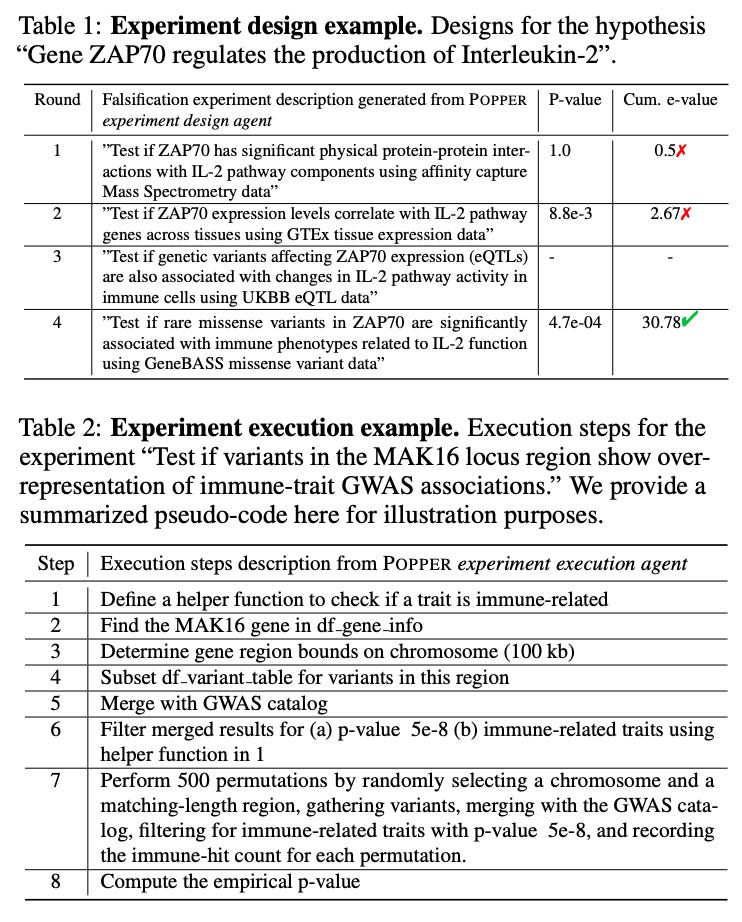
Automated Hypothesis Validation with Agentic Sequential Falsifications [Huang et al., aRxiv, February 2025]
Teams at Stanford and Harvard asked themselves “How can we rigorously validate free-form hypotheses at scale?”
The authors developed an agentic framework for rigorous automated validation of free-form hypotheses named POPPER.
Based on Karl Popper’s principle of falsification, POPPER validates a hypothesis using LLM agents that design and execute falsification experiments targeting its measurable implications. POPPER challenges hypotheses by sequentially testing their measurable implications through diverse experiments, ranging from data analysis and simulations to real-world experiments and interventions.
POPPER employs two agents: the Experiment Design Agent which leverages reasoning capabilities and domain knowledge to identify a measurable implication of the main hypothesis and designs a falsification experiment; and the Experiment Execution Agent which implements the experiments, which could involve data collection, simulations, statistical analyses or real-world experiments. This all leads to a p-value that summarizes the outcome of the experiment and then converted into e-values, enabling the aggregation of cumulative evidence.
They demonstrated the framework on six domains including biology, economics and sociology that effectively controls Type-I error rate (falsely rejecting true null hypothesis) and shows equal performance to human scientists (in the biology field), whilst reducing time to validation 10-fold.
Genome modelling and design across all domains of life with Evo 2 [Brixi et al. 2025]
Brian Hie’s lab at the Arc Institute has released Evo 2, their latest foundation model trained on genomes from across the evolutionary tree. This represents a major advance on their original Evo model which was only trained on prokaryotic genomes. The full-scale Evo 2 model includes an incredible 40B parameters and 9.3 trillion tokens.
The authors have evaluated Evo 2 in a variety of ways:
Predicting mutational effects on molecular function or organismal fitness. These tests show that Evo 2 reflects the expected importance of different genetic alterations.
Zero-shot variant effect prediction. Evo 2 outperforms other models when predicting non-coding variant effects and performs comparably to existing models for coding variant effect prediction.
Beyond benchmarking the model, the authors show that the latent representations of Evo 2 capture a broad spectrum of biologically relevant signals including protein secondary structure, regulatory elements and exon/intron boundaries. Using the semantic mining approach developed in Merchant et al., 2024 the authors use Evo 2 for various sequence completion tasks. These experiments show that Evo 2 can generate sequences including coding and non-coding elements resembling organelle, prokaryotic and eukaryotic genomes. Finally, the authors couple Evo 2 to sequence-to-function models such as Enformer and Borzoi to design sequences where the location and length of chromatin-accessible regions is varied. This is used to encode Morse code messages into DNA reading “ARC” and “EVO2”. Overall, this paper lays the foundation for de novo genome design and enables many exciting applications in synthetic biology.
Notable deals
Altitude Lab has launched a pre-seed venture fund, anchored by Recursion CEO Chris Gibson, to support early-stage biotech startups affected by recent federal funding shifts. The Altitude Lab Fund aims to bridge the funding gap for SBIR-reviewed companies, providing $100,000–$250,000 in investment, 12 months of lab space, and mentorship through Altitude Lab’s accelerator.
Epitopea forms research and licensing agreement with Merck. The pair will leverage Epitopea’s CryptoMap which identifies tumor-specific antigens derived from so-called “junk DNA,” unlocking new targets for immunotherapy. The collaboration aims to develop a portfolio of novel cancer treatments, with Epitopea potentially earning up to $300 million per program.
Isomorphic Labs and Novartis expand collaboration, adding three additional research programs to the original agreement. It remains unclear whether this expansion is due to the initial programs reaching key milestones, challenges or insights from the early efforts prompting a shift in focus, or a strategic reprioritization of research priorities.
Atrandi Biosciences, a Lithuania-based biotech, has secured $25M from Lux Capital to expand into the U.S. in 2025. The company’s Semi-Permeable Capsule (SPC) technology enables high-throughput single-cell analysis, offering a unique alternative to industry giants like 10x Genomics and Illumina. Lux investor David Yang discovered Atrandi through researchers already using the tool, highlighting strong user validation before investment.
Newleos Therapeutics has emerged with $93.5 million in Series A funding, led by Goldman Sachs Alternatives, to advance a portfolio of licensed CNS drugs from Roche. The startup aims to generate proof-of-concept data over the next two and a half years for treatments targeting generalized and social anxiety disorders, as well as substance use disorders. Led by Longwood Fund’s David Donabedian, the company licensed four clinical-stage assets, including NTX-1955, a selective GABAA modulator designed to offer a safer alternative to benzodiazepines.
What we liked on socials channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here.