BioByte 103: drug discovery's DeepSeek moment, microfluidics and deep learning for mapping proteins, a novel means of metabolite detection, and evaluating Deep Research with a tox task
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
An Evaluation of "Deep Research" Performance [Derek Lowe, Feb 2025]
OpenAI’s new Deep Research tool is designed to handle multi-step research tasks, synthesizing information from across the web. Derek Lowe tested it on a task assessing thalidomide’s toxicity across species and isomers. It is clear the tool has strong potential but also faces challenges that need to be addressed.
Where It Can Improve:
Prioritizing recent research
Deep Research pulls from a broad range of sources, but it does not yet differentiate well between older hypotheses and more recent discoveries. In scientific fields where knowledge evolves rapidly, ensuring that newer findings take precedence will be key to improving accuracy.
Balancing breadth and precision
The tool aims to be comprehensive, but this sometimes results in an overwhelming amount of information. When explaining why thalidomide affects species differently, it included both outdated metabolic hypotheses and the now well-established cereblon mechanism. With better filtering, it could focus on the most relevant and up-to-date explanations.
Understanding technical terms in context
Deep Research confused thalidomide’s chiral stability (racemization) with its metabolic stability (half-life and breakdown), which could lead to misunderstandings. Improving its ability to distinguish between technical definitions will make its reports more reliable.
Recognizing the bigger picture
The tool stayed within the requested scope but missed a major contextual point. The discovery that thalidomide binds to cereblon did not just explain its toxicity, it led to the entire field of targeted protein degradation. A human researcher would naturally highlight this, and helping the model make those connections will significantly enhance its usefulness.
The Drug Industry Is Having Its Own DeepSeek Moment [David Wainer, WSJ, February 2024]
DeepSeek sent shockwaves through the markets and dominated the news when it announced the release of its R1 model three weeks ago. Though perhaps not making as big of headlines, China’s rise in competitiveness in tech has been powerfully mirrored in biotech as well. Wainer cites this trend as beginning last fall when Summit Therapeutics’ drug candidate—licensed from Chinese biotech, Akeso—beat out Keytruda, Merck’s long-reigning immunotherapy star, in a ‘head-to-head lung-cancer trial.’ While not yet even approved for market, this news resulted in a multi-billion dollar boost to Summit’s market cap. Instances such as this are becoming increasingly frequent as US pharmaceutical companies are extending their search for drug candidates beyond their traditional domestic shopping grounds of US-based biotechs. This is notably exemplified within the highly competitive and lucrative GLP-1 space as of late, especially as the hunt for an oral GLP-1 heats up. In efforts to keep up with Eli Lilly’s and Novartis’ injectables, Merck and AstraZeneca are both highlighted as turning to China. While Merck had been initially prospecting Viking Therapeutics—a US company with a $3.7 billion market value—for their oral GLP-1, they ultimately went with China’s Hansoh Pharma for a deal of only $112 million upfront and still only up to $1.9 billion in milestone payments. AstraZeneca made a similar move a year preceding Merck’s. And, by basic economic principles, why wouldn’t they? As Wainer so succinctly puts it, “Why spend $10 billion acquiring a U.S. biotech with a mid-stage drug when a similar molecule can be licensed from China for a fraction of the price?”
While this trend represents a victory for Big Pharma and patients alike—who have only to gain from the increased competitiveness—it confers a challenge to US biotechs and the venture firms that back them. Building a successful biotech is generally a long and expensive journey, but this growing pressure from China suggests an increasing need to find ways to cut costs and increase efficiency in the process. Much like Deepseek’s origin, Chinese biotechs are described as ‘scrappier,’ utilizing a workforce that can work faster for less and, moreover, that is highly skilled, in no small part due to the many scientists trained in the US returning to China in the past decade.
With pharma transactions in China having leapt a whopping 25% in the past five years and the S&P biotech ETF remaining ultimately flat for the past two (whilst the S&P 500, largely concentrated in tech, has climbed 48%), there is major market signaling for US biotechs to adapt or get out-competed. Still, it is important to note that much of China’s edge is due to minor improvements on existing drug formulations rather than pioneering discoveries. It will be fascinating to see how US drug discovery companies reconcile leading innovation with pursuing lower costs and higher efficiency.
Papers
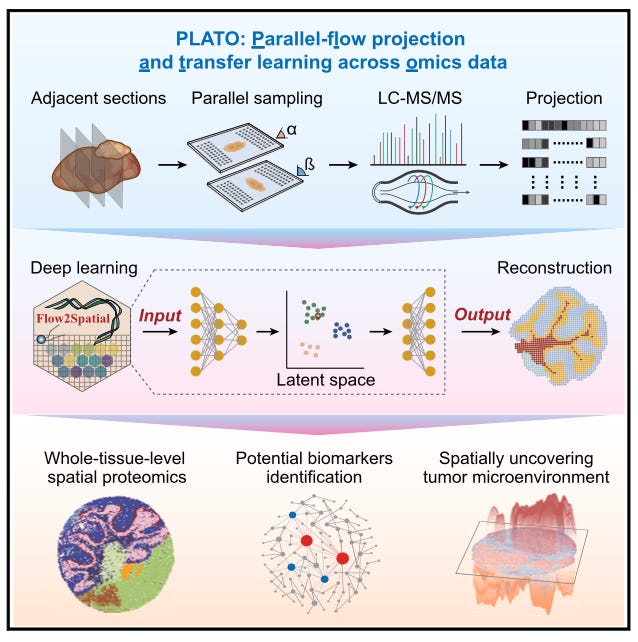
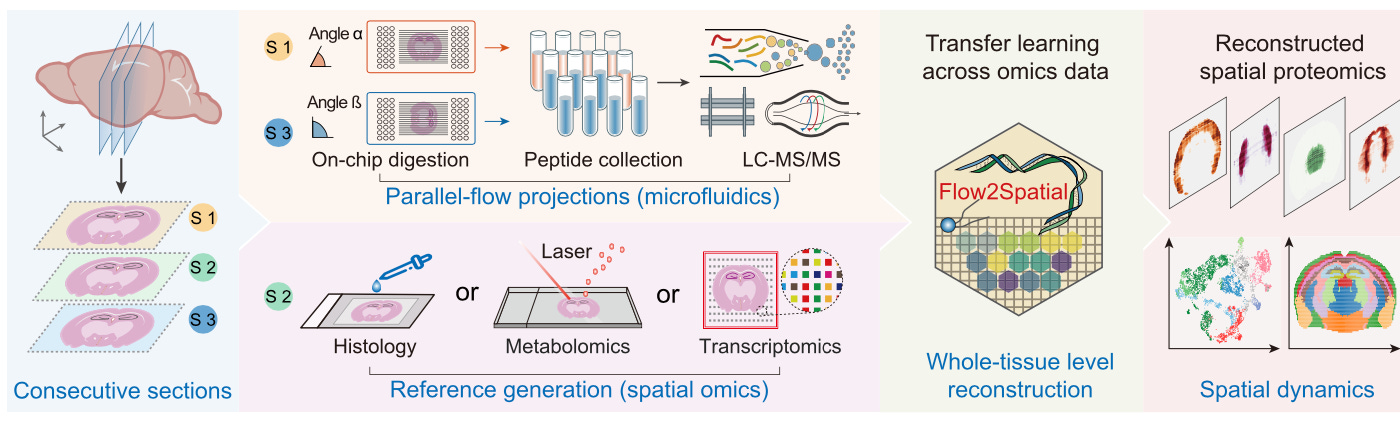
High-resolution spatially resolved proteomics of complex tissues based on microfluidics and transfer learning [Hu et al., Cell, 2025]
High-resolution, high-throughput protein mapping across entire tissues remains a significant challenge. To solve this, the authors, from the University of Chinese Academy of Sciences, Beijing Institute of Lifeomics and Peking University Cancer Hospital present a new framework which combines microfluidics with deep learning.
The platform is called PLATO or parallel-flow projection and transfer learning across omics data. Its workflow begins by sectioning three tissue slices. The middle is used to generate reference transcriptomic or metabolomic data through histological staining or spatial omics. The first and last slices are processed using microfluidics-based proteomic profiling. After digestion, peptides are then collected for LC-MS/MS analysis. To reconstruct spatial protein distributions they used a transfer learning model called Flow2Spatial, trained on omics data (such as the one from the reference slice) to predict predict protein distribution from the parallel-flow data.
PLATO was able to identify more than 2,000 protein groups in a single run from a mouse cerebellum sample. At a resolution of 25 microns, they uncovered protein dynamics associated with disease states, which revealed distinct tumor subtypes and identified dysregulated proteins.
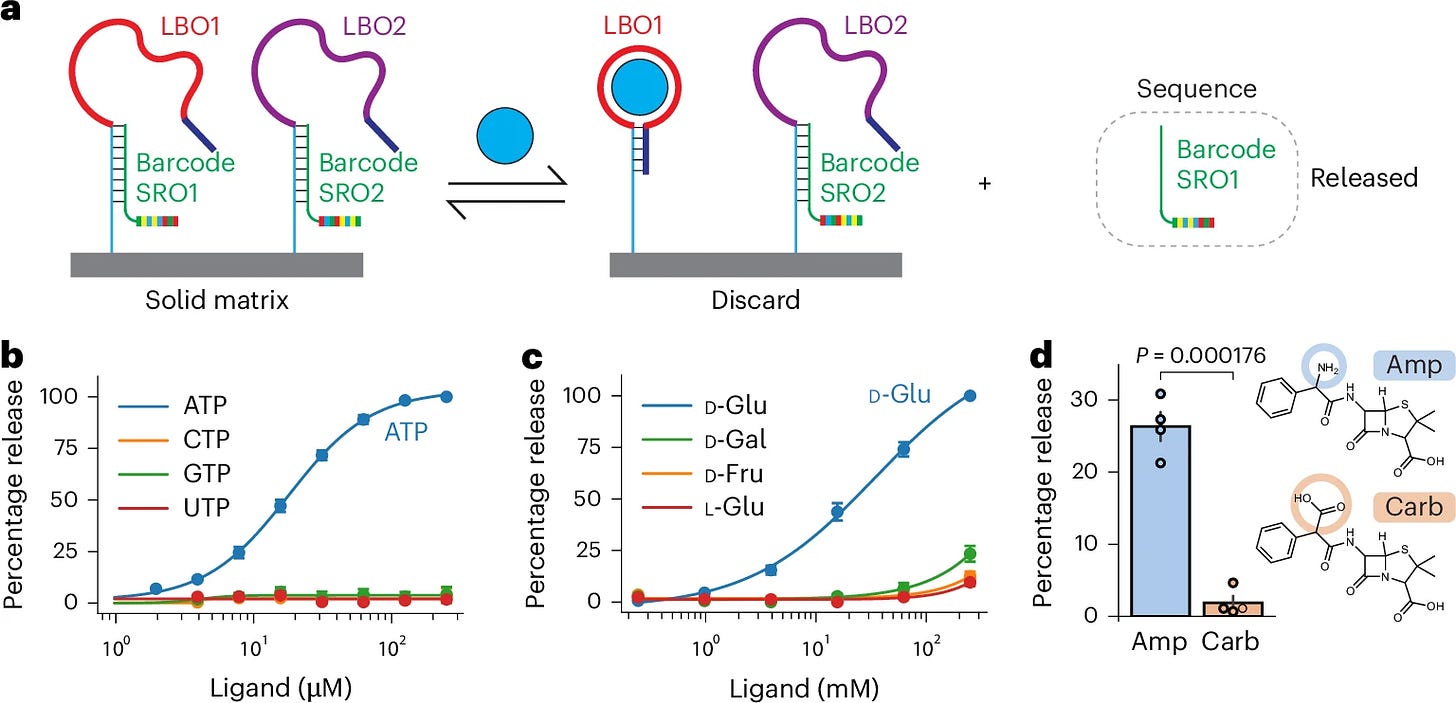
Quantifying metabolites using structure-switching aptamers coupled to sequencing. [Tan & Fraser, Nature Biotechnology, 2025]
Detecting small molecules like metabolites is challenging because unlike nucleic acids, they lack modular building blocks that can be read out using sequencing. To overcome this, June Tan and Andrew Fraser from University of Toronto have developed small-molecule sequencing (smol-seq) - a clever approach that converts the presence of a metabolite into a sequencing readout.
Central to their approach are structure-switching aptamers (SSAs). SSAs are small nucleic acid sensors that undergo a major conformational change when binding their target molecule. In this paper, these SSAs have been adapted so that when their target binds a short release oligo (SRO) is displaced from the longer ligand-binding oligo (LBO). Each SRO contains a unique barcode that corresponds to a specific metabolite, allowing its presence to be detected using sequencing.
Smol-seq can detect 100s of metabolites in parallel with high specificity and could be integrated with other sequencing workflows for multi-omics applications. Furthermore, the approach could be adapted to do single-cell metabolomics or even spatial metabolomics. In future, the authors also plan to expand their SSA library to detect more diverse mixtures of metabolites with the potential to transform how we detect metabolites and small molecule therapeutics.
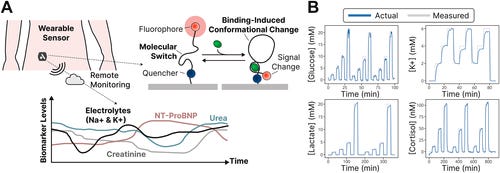
Continuous molecular monitoring for precision heart failure care [Alex Yoshikawa, JACC: Basic to Translational Science, Feb 2025]
Heart failure (HF) remains one of the biggest challenges in healthcare, with high hospital readmission rates driving up costs and worsening patient outcomes. Despite advancements in treatment and monitoring, one in four heart failure patients is readmitted within 30 days, and nearly half return within six months. The problem is that current methods—like periodic checkups, symptom tracking, and lab tests—often miss the early warning signs of deterioration.
To change that, Alex and his team have developed the HF Monitor, a wearable biosensor designed to transform heart failure management outside the hospital. Instead of waiting for symptoms to appear, this device continuously tracks real-time molecular data, allowing doctors to detect problems before they escalate.
The HF Monitor is a small, non-invasive device that sticks to the abdomen, much like a continuous glucose monitor. It features a flexible probe that collects and analyzes key heart failure biomarkers from interstitial fluid under the skin. These biomarkers provide a far more accurate picture of a patient’s heart and kidney function than simply tracking weight or blood pressure.
Specifically, the device measures:
NT-proBNP – A crucial biomarker that indicates the severity of heart failure.
Sodium & potassium levels – Essential for heart function and medication balance.
Creatinine & urea – Markers of kidney function, which is closely linked to heart failure progression.
What sets the HF Monitor apart is its cutting-edge molecular switch technology. Unlike traditional biosensors that rely on naturally occurring enzymes (which can only detect a limited set of molecules), this device uses engineered aptamer-based switches that can be customized to detect a wide range of biomarkers in real time. These molecular switches change their fluorescence intensity when they bind to specific biomolecules, allowing for continuous, real-time measurement of critical HF markers.
This is a major breakthrough because it allows multiple biomarkers to be tracked simultaneously in a single device, eliminating the need for separate tests and monitors. Even more powerful, the real-time data can be paired with AI-driven analytics, helping doctors identify trends, predict worsening conditions, and intervene before a crisis occurs. The same continuous molecular monitoring platform can be adapted for chronic kidney disease, drug monitoring, critical care, general wellness, and even clinical trials, opening the door for real-time, data-driven healthcare across multiple conditions.
Notable deals
Latent Labs secures $50M in funding to realize the potential of AI-powered, programmable biology. The raise includes $40M of fresh Series A funding from Radical Ventures and Sofinnova Partners. Simon Kohl, CEO and Founder, was previously a lead researcher in the AlphaFold2 team.
San Francisco-based Junevity has secured $10 million in seed funding to develop siRNA therapies that silence transcription factors linked to aging and disease. Backed by Goldcrest Capital and Godfrey Capital, the startup leverages machine learning to map how gene expression differs between young and old cells, identifying key transcription factors driving disease progression. Junevity’s lead program is an insulin sensitizer for type 2 diabetes, designed to control blood sugar without the weight gain and heart risks associated with existing treatments. It was co-founded by Janine Sengstack and based on research from UCSF’s Hao Li lab.
Terrain Biosciences has launched to design and manufacture AI-optimized RNA, solving key challenges in stability, immune evasion, and scalability. Founded by ex-Tome Biosciences team members Omar Abudayyeh and Jonathan Gootenberg, and backed by $9 million from Magnetic Ventures, Bruker Corporation, and industry veterans, Terrain provides biotech startups with high-quality RNA to eliminate the need for costly in-house production. CEO Chetan Tadvalkar emphasizes a lean, revenue-driven approach over massive funding rounds, with a Series A planned later this year. By integrating AI-driven design with rapid, scalable manufacturing, Terrain cuts RNA turnaround times by up to sixfold, allowing researchers to focus on discovery rather than logistics. Much like NVIDIA for chips or TSMC for manufacturing, Terrain aims to be the backbone of RNA innovation, accelerating the future of medicine.
AbbVie inks T-Cell engager deal with Xilio for $42M upfront + $10M equity investment in a “multi-program” collaboration. T-cell engagers bind to both T-cells and a target cell, bringing them into proximity for each other for therapeutic action. Xilio’s technology focuses on a concept of ‘masking’ which aims to limit therapeutic activity in the periphery.
ExpressionEdits partners with Boehringer Ingelheim to enhance gene therapies using its AI-driven Genetic Syntax Engine, optimizing introns to boost protein expression without altering genes. Boehringer gains access to this technology for undisclosed targets, with potential milestone payments.
Bioluminescence Ventures shuts down. The firm had raised half a billion dollars to invest into new biotechs, but due to a limited number of LPs and the seemingly sudden withdrawal of a primary investor in late January, it was forced to close down less than a mere two years after founding. With an already promising portfolio, we’re sorry to see them go.
In case you missed it
The LongBio Report 2025 by Healthspan Capital
What we listened to
What we liked on socials channels
Bonus for all our fellow em dash (and AI) fans:
Events
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here.