BioByte 094: industry consortium for benchmarking, evaluating protein competitions, multimodal atlas for Alzheimer's, automated organelle tracking, promise of a new HAE treatment
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
A call for an industry-led initiative to critically assess machine learning for real-world drug discovery [Nature Machine Intelligence, Oct 2024]
Benchmarking in drug discovery is severely lacking. A new cross-industry initiative called Polaris is focused on bridging the gap between perceived progress and real-world impact in machine learning for drug discovery, particularly in small-molecule predictive modeling. Unlike other fields with abundant fit-for-purpose datasets, drug discovery's data is heterogeneous, noisy, and expensive to generate, making reliable ML benchmarking crucial but challenging.
Polaris brings together industry experts to address these unique challenges through cross-industry collaboration. The first steering committee—made up of members from various pharma and drug discovery companies like Merck, Relay Tx, and Valence Labs, has highlighted the need for robust benchmarking practices to evaluate ML methods in real-world contexts. Their collective effort will develop domain-appropriate datasets, guidelines, and open-source tools to standardize method evaluation and comparison, ensuring reproducibility and progress in drug discovery.
The initiative prioritizes three pillars:
Curating benchmark datasets representative of common drug discovery tasks.
Establishing best practices for data curation, method evaluation, and statistical comparison.
Creating open-source tools to simplify adopting these practices.
By fostering interdisciplinary collaboration and real-world benchmarking, Polaris aims to accelerate impactful ML research and advance drug discovery. Be on the lookout for their first preprint comparing methods and best practices.
AI has dreamt up a blizzard of new proteins. Do any of them actually work? [Ewen Callaway, Nature, October 2024]
With the recent the recent Nobel Prize in Chemistry awarded to David Baker, Demis Hassabis and John Jumper for their work on protein design (David Baker) and structure prediction (Hassabis and Jumper), it is difficult to deny that computational protein design is going to shape the future of protein-based research. Since it is such a new field, organizations across the globe—like Liberum Bio, Rosetta Commons, and Adaptyv Bio—have begun hosting competitions that encourage competitors to further develop design tools. The challenges range from redesigning an existing enzyme used in protein purification to building a new protein that can be used in T-cell cancer therapy.
These competitive events are largely inspired by the Critical Assessment of Structure Prediction (CASP), which has been around since 1994 and which ultimately facilitated the creation of AlphaFold and AlphaFold2. The historic success of such structural prediction competitions has caused many to look favorably upon their implementation for novel protein design, as they have the propensity to serve as significant instigators for acceleration within the field. Proponents also tout the accessibility of the competitions as a way to involve even those without formal biology training or access to computational clusters. There is hope that the competitions will also help develop and strengthen the community.
Nonetheless, these contests are not without their deficiencies. Too many novel protein designs can lead to confusion over the validity of such a myriad of diverse approaches, as the verification process in the lab is far slower. Given the multitude of protein applications, there are also concerns that the approaches used to design one protein will not be translatable to another. Furthermore, the benefit to the field largely hinges on participants sharing their methods to facilitate communal learning. This latter conundrum has fortunately thus far not proven to be a significant issue, however, as most competitions require or heavily encourage participants to describe their approach as part of their entry.
Although it is early, these competitions appear to be here to stay, with Adaptyv Bio having recently launched a second competition that builds on the first. With more accessible, powerful computational tools that lower barriers to entry–such as later versions of AlphaFold and EvolutionaryScale–these contests will continue to attract the brightest minds to the space. Still, even as the competitors produce countless proteins to tackle some of the world's most daunting biological problems, only time and rigorous testing will tell if these designs hold out as hits in the lab.
Machines of Loving Grace [Dario Amodei, Oct 2024]
Dario Amodei, cofounder of Anthropic, put out a new essay presenting an optimistic look at how AI could dramatically improve many areas of human life. While he acknowledges the risks, the focus is on the positive changes AI could bring is a refreshing take. He highlights five main areas where AI could have the biggest impact with two of the five notably being rooted in biotech and health:
Health and Biology: AI could revolutionize medical research and treatment, speeding up the discovery of cures for diseases, creating personalized treatments, and extending human life. Amodei believes AI could accomplish 50 to 100 years' worth of medical advancements in just 5 to 10 years, tackling major health challenges like cancer, Alzheimer's, and genetic conditions. He specifically thinks AI will double the human lifespan and drive personalized treatments.
Mental Health and Neuroscience: AI could offer breakthroughs in understanding the brain, leading to better treatments for mental health issues like depression, addiction, and schizophrenia. It could also help improve overall mental well-being, allowing people to enhance their cognitive abilities and emotional health on a day to day basis.
Economic Development and Poverty: AI has the potential to accelerate economic growth, especially in poorer regions. By improving health, optimizing supply chains, and introducing advanced technologies, AI could help close the gap between wealthy and developing countries, boosting productivity and lifting people out of poverty.
Governance and Peace: AI could help make smarter decisions in governance, reduce conflict, and fight corruption. By improving systems for diplomacy and public policy, AI could contribute to more stable societies and reduce human suffering caused by governance issues.
Work and Purpose: AI could change the nature of work, taking over routine tasks and giving people more time to focus on creative and meaningful activities. While there are concerns about job loss, AI could also open up opportunities for people to pursue more fulfilling careers and ways of living.
Amodei remains cautious, emphasizing the need to carefully manage AI’s power to avoid making inequalities worse or creating new problems and maintains the need for human oversight and involvement. Still, he is hopeful that if AI is handled responsibly, it could bring about a "compressed 21st century," where progress happens at an accelerated pace.
Genome Editing for Hereditary Angioedema Promises a Life Free from Disease Burden [Dirk Haussecker, RNAi therapeutics, 2024]
Intellia Therapeutics is close to releasing phase II data for NTLA-2002, a novel one-time treatment for hereditary angioedema (HAE). This rare genetic disorder causes unpredictable and potentially life-threatening swelling attacks, significantly impacting patients' quality of life. Current treatments, while helpful, often fall short in providing complete protection and require regular administration, leading to poor quality of life and high healthcare costs.
NTLA-2002 employs CRISPR-based genome editing to permanently disrupt the KLB1 gene in liver cells, which codes for plasma prekallikrein (PKK), a crucial component in the inflammatory system responsible for HAE attacks. The treatment, delivered via lipid nanoparticles, has shown promising results in phase I trials. All ten subjects became essentially attack-free following a single administration, with minimal side effects limited to the time of treatment.
The upcoming phase II data is expected to provide more comprehensive information on dosing, safety, and efficacy. Based on phase I results, the company is likely to select a 50mg dose for further testing, which demonstrated an 88% reduction in plasma PKK levels. This reduction is significantly above the threshold (~45%) where HAE attack rates significantly decrease. If successful, NTLA-2002 could offer patients significant relief from HAE, potentially eliminating the need for ongoing treatments and associated costs.
While the potential price tag of NTLA-2002 may be high, estimated at $2-3 million per patient, it could prove cost-effective in the long run compared to current treatments that often exceed $500,000 annually. Although costly, NTLA-2002 represents a significant advancement in medical innovation for HAE patients.
Papers
Integrated multimodal cell atlas of Alzheimer’s disease [Gabitto et al., Nature Neuro, Oct 2024]
The Allen Institute for Brain Science published this week in Nature Neuroscience a multimodal cell atlas of Alzheimer's disease (AD) progression in the human brain. While the accumulation of amyloid plaques and tau tangles in AD is well-characterized, the specific cell types affected and molecular changes occurring over the course of disease progression remain poorly understood.
Using postmortem brain tissue from 84 donors spanning the range of AD pathology using single-nucleus RNA sequencing, ATAC-seq (epigenetic changes), spatial transcriptomics, and quantitative neuropathology two major epochs of AD disease progression were discovered: 1) an early phase with slowly increasing pathology, characterized by microglial and astrocyte activation, loss of specific somatostatin interneurons in superficial cortical layers, and oligodendrocyte changes; and 2) a late phase with exponential increases in amyloid and tau pathology, featuring broader neuronal loss (especially layer 2/3 intratelencephalic excitatory and parvalbumin/VIP inhibitory neurons), continued glial activation, and more extensive cellular dysfunction across multiple cell types. This biphasic progression highlights the distinct cellular and molecular changes occurring at different stages of AD pathology.
These findings highlight the importance of early therapeutic intervention that target specific cell types involved in the initial epoch of disease progress (e.g., microglial-targeting drugs) in order to prevent transition to the more aggressive late phase of the disease characterized by increases in amyloid and tau pathology.
Nellie: Automated organelle segmentation, tracking, and hierarchical feature extraction in 2D/3D live-cell microscopy [Lefebvre et al., arXiv, 2024]
Image analysis of dynamic organelles remains a challenge due to the “limitations inherent to microscopy such as acquisition speed, the diffraction limit, and tradeoffs between signal and phototoxicity, pose significant challenges in extracting this information”. In order to provide detailed extraction of spatial and temporal features at multiple organellar scales, the authors developed Nellie. Nellie is a computational pipeline that automates the segmentation, tracking, and feature extraction of organelles, eliminating user bias and saving time.
In the paper, Nellie demonstrates applicability across a wide range of applications in cellular biology: 1) in its ability to capture extensive metrics in both Golgi and mitochondria by using object and branch-based morphology and motility features to train random forest models and predict organelle type, 2) it was able to construct multi-level mesh-like graph networks of mitochondria to then train a first-of-its-kind organelle-based graph autoencoder which the authors claim it is reminiscent of cell painting methods (treatment clustering and comparisons) and GraphCast’s weather prediction (local morphology and motility predictions) and 3) detected meaningful differences in endoplasmic reticulum networks between cells while maintaining consistency across temporal frames within the same cell.
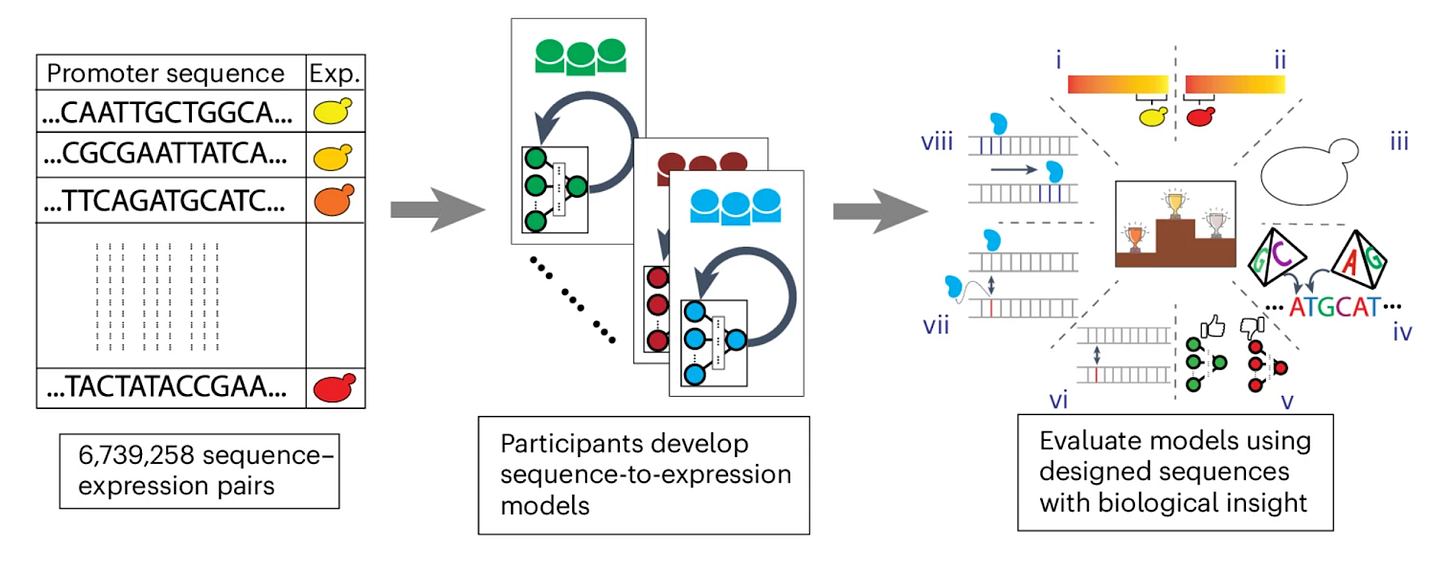
A community effort to optimize sequence-based deep learning models of gene regulation [Rafi et al., Nature Biotechnology, October 2024]
Researchers organized a community challenge to develop better machine learning models for predicting gene expression from DNA sequences. The DREAM Challenge provided participants with a large dataset of random promoter sequences and corresponding expression levels in yeast. Teams competed to create models that could most accurately predict expression from sequence alone.
The top-performing models outperformed previous state-of-the-art approaches and generalized well to other species and experimental setups. Convolutional neural networks dominated, though recurrent and attention-based models also performed well. Innovative training strategies proved as important as model architecture in improving performance. A modular "Prix Fixe" framework allowed mixing and matching of model components, leading to further improvements.
Surprisingly, simpler models with fewer parameters could match or exceed more complex models if optimized well. This work provides a roadmap for continued improvement of genomics models through standardized datasets, evaluation metrics, and modular frameworks. The improved models can help researchers better understand how genetic variants impact gene expression and identify important regulatory elements in the genome. Ultimately, more accurate models of gene regulation can better support research into the genetic basis of diseases and aid in developing new therapies.
Notable deals
Shift Bioscience raises $16M to ‘reverse aging’ with an AI cell simulation
The Cambridge, UK biotech is collecting data on how the activation of different genes affect a cell’s aging process. By combining this with machine learning, it is hoping to develop a ‘cell simulation’ which it can use to probe with therapeutics that could reverse the ‘age’ of a cell whilst maintaining its development. Investors include BGF, F-Prime and Kindred Capital.
Pfizer partners with molecular glue start-up Triana for $49M upfront
Forbion raises $1.3B growth and $970M venture biotech funds aimed at 15 companies
What we liked on socials channels
How to Choose a Good Scientific Problem
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone