BioByte 087: Push for biodiversity data, Recursion's Big Moment, Conditional Generation of Molecules, Graph AI Applied, Evolved Hackathon, and more
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
Hope everyone is surviving the worst form of seasonal depression—the transition away from summer. Thanks for bearing with us as we took a little post summit break, we’re now back with your weekly reads!
What we read
Blogs
Nature-rich nations push for biodata payout [FT, August 2024]
The UN is working on a fund to pay stewards of biodiversity, mainly communities in lower-income countries, for discoveries made with genetic data from their ecosystems. This mechanism was established in the 2022 UN Convention on Biological Diversity. How it will be funded and governed will be determined at the COP16 Summit.
Pharmaceutical, food and agritech companies have been using biodiversity hotspots to mine for useful molecules and genetic data to use in their products for decades. As an example, the author mentions how Phytofarm licensed the active ingredient from an African cactus that caused appetite suppression without compensating the indigenous people that discovered it.
If the fund comes to fruition it would raise billions. The strategy to raise the funds is to raise public pressure and ambition for industries to make voluntary contributions. The company Basecamp Research, featured in the article, works with governments to establish partnerships. If the company uses their data for a product or service, the respective governments will receive royalties.
Recursion nears ‘moment of truth’ with first key data, kicking off 18-month flurry of readouts [Andrew Dunn & Max Gelman, Endpoints, August 2024]
Recursion is gearing up for a big moment in the world of AI-powered drug discovery. They're about to drop their first major clinical trial readout, kicking off a whirlwind 18 months where they'll share data from 10 different trials...an early goal of the company many thought too ambitious at the time.
Interestingly, their lead drug wasn't even created by AI—it's a repurposed compound they're testing on a rare brain condition; however, the company’s pipeline includes several AI-designed drugs that could serve as tremendous proof of concept for the world of AI-first biotech.
These trial results aren't just big for Recursion—they could shape the future of AI in drug development. If it works, it could be desperately needed validation for the field. If not, well, it might send some investors running for the hills (just kidding). While it is now indisputable across biotech that AI does have a large role to play in drug and trial design, the biotech world is watching closely to see if Recursion can turn all that AI hype into safe and effective therapies for diseases with high unmet need. Though we caution that the expected outcome for most drug development is, unfortunately, failure—putting too much weight on the results of any one readout may change near-term market sentiment but should not reflect the state of the industry at-large.
Predicting New Small Molecule Binders [Derek Lowe, In the Pipeline, August 2024]
The results of Leash Bio’s Kaggle competition predicting new small molecule binders are out and the results are…”well about what you would expect”. As a quick recap, the competition made a dataset of DNA-encoded library screens on 133 million chemical species (mostly trisubstituted traizines, which is a standard combinatorial chemistry motif) available to any data scientist or researcher who wanted to take a shot at generating compound-binding models. Over 2,000 participants took a shot at it with no single team producing great results. There are many reasons this could happen. For one, it’s unreasonable to expect strong zero-shot learning from such a setup. It’s also unclear what type of techniques were used as it was an open Kaggle competition. Other explanations are that the dataset was not dependent on orderable molecules from existing catalogs like other studies have had or the type of data generated by DEL screens is unsatisfactory, which Lowe comments is unlikely to be the case. While this particular competition was underwhelming in results, we’re all for more companies open-sourcing their datasets and contributing to benchmarking efforts!
Papers
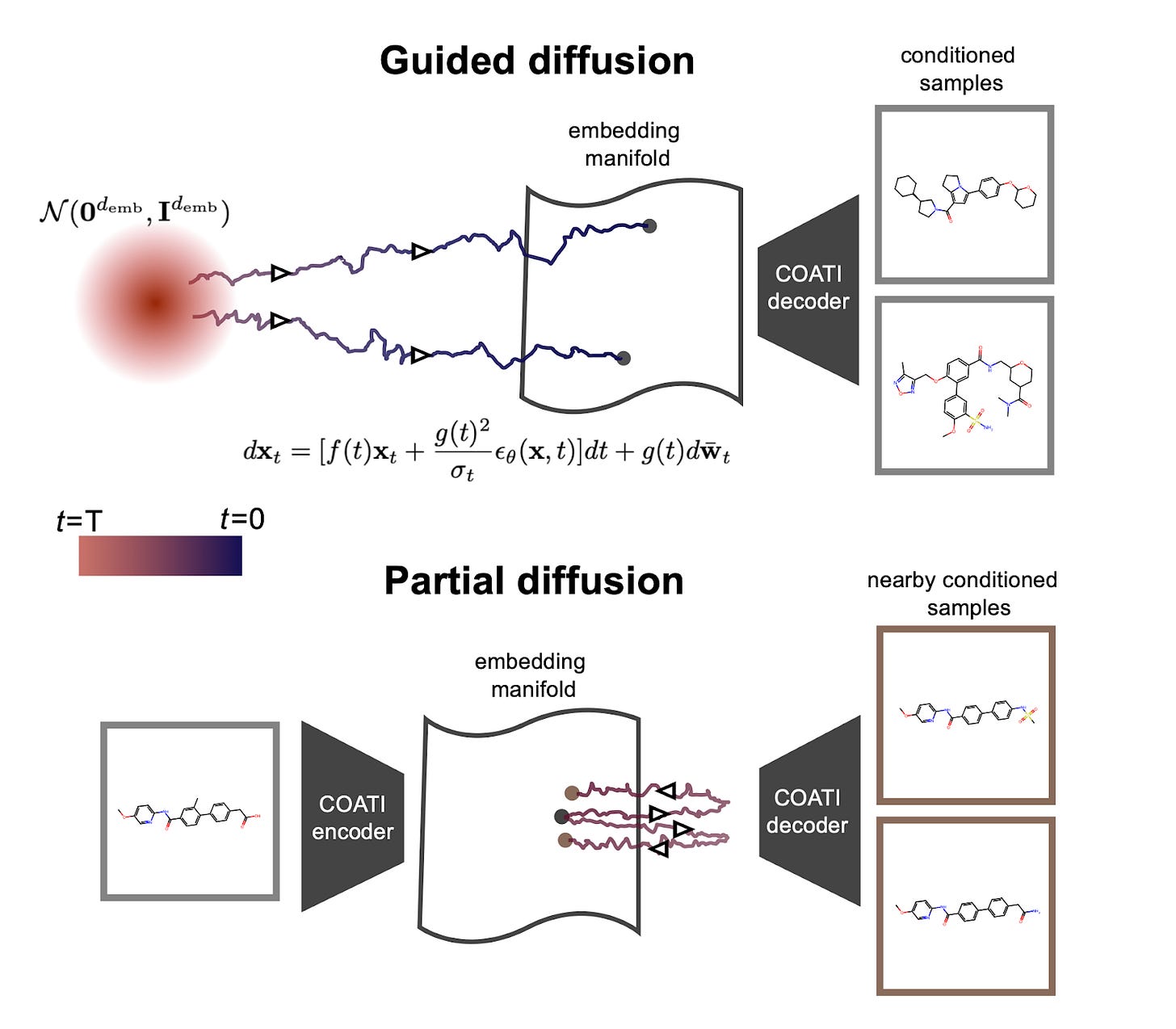
Latent Diffusion for Conditional Generation of Molecules [Kaufman et al., BioRxiv, August 2024]
Terray Therapeutics published a preprint this week detailing a new generative algorithm for the conditional generation of small molecules optimized for multiple therapeutically-relevant properties. Unlike other approaches where diffusion models are applied to molecular structures directly, COATI-LDM generates embeddings from molecular structures, and then applies diffusion models on these latent vectors. This approach offers several advantages:
Flexibility: COATI-LDM can easily incorporate various molecular properties and optimize for multiple objectives simultaneously.
Efficiency: The method performs well even with limited training data, a common constraint in drug discovery.
Control: Researchers can guide the generation process to produce molecules with specific desired properties
The team showed that the model could generate molecules with tightly controlled properties, such as lipophilicity (how well a molecule dissolves in fats) and binding affinity to a specific protein target. COATI-LDM outperformed traditional genetic algorithm approaches in generating diverse sets of potential drug candidates. The team even incorporated a medicinal chemist preference score, allowing the model to generate molecules that align with expert intuition about what makes a good drug candidate. Finally, a "partial diffusion" technique was also developed that allows for fine-tuning existing molecules, a crucial capability for lead optimization in drug discovery.
Multiplexed single-cell characterization of alternative polyadenylation regulators [Kowalski et al., Cell, August 2024]
Rahul Satija and Anshul Kundaje’s labs teamed up to develop a new method called CPA-Perturb-seq, a high-throughput method that combines CRISPR genetic perturbation with single-cell RNA sequencing to study how perturbations of 42 known cleavage and polyadenylation (CPA) regulators affect polyA site usage across the transcriptome. Alternative polyadenylation (APA) is a post-transcriptional regulatory mechanism that generates diversity in mRNA transcripts. Most mammalian genes have multiple polyadenylation sites, allowing a single gene to produce multiple mRNA isoforms. While APA is known to play roles in various biological processes, the regulatory mechanisms governing polyadenylation site choice are not fully understood. Understanding the regulation of APA is crucial because it can affect multiple aspects of RNA biology, including transcript stability, localization, and protein production.
The team identified distinct modules of co-regulated polyA sites, each responsive to perturbation of different regulators or subcomplexes. For intronic polyA sites, different sets of sites were identified that are uniquely sensitive to perturbation of factors regulating distinct components of the nuclear RNA life cycle, including elongation, splicing, termination, and surveillance. The researchers developed a deep neural network model to predict perturbation responses from RNA sequences, revealing interactions between regulatory complexes. CPA-Perturb-seq will be a powerful tool for studying mechanisms of post-transcriptional regulation, leading to a deeper understanding of RNA biology.
Spatially clustered type I interferon responses at injury borderzones [Ninh et al., Nature, 2024]
Heart attacks (myocardial infarctions, MIs) have been increasing in frequency, and the sequelae that follow make it the most common cause of death worldwide. A serious complication of MIs is a myocardial rupture, which is associated with a >50% chance of death. Myocardial ruptures happen at the border between injured and damaged cardiac tissue, and was believed to be a confluence of inflammation, poor wound healing, and mechanical stresses that led to failure of compromised tissue. However, the molecular mechanisms underlying the pathogenesis of ruptures has remained poorly understood.
Here, the authors analyzed the infarcted hearts of human and mice hearts using genome-wide spatial transcriptomics and single-cell, spatially resolved RNA and DNA multiplexed error robust fluorescence in situ hybridization (MERFISH). The authors consistently found a gene-expression signature in the borderzone that suggested activation of type 1 interferon signaling (a component of innate immunity), and was distinct from other markers of innate activation found in other areas of cardiac damage. In particular, they observed clusters of interferon activation that spanned a short layer of cells that was present early after an MI and. Several cell types were involved in this cluster, including cardiomyocytes, fibroblasts, endothelial cells, and various immune cells.
By generating mice with cell-type-specific deletion of the interferon regulator gene Irf3 in cardiomyocytes, fibroblasts, macrophages, neutrophils, and endothelial cells, the researchers determined that cardiomyocytes are the dominant initiators of the interferon response in the borderzone. Additionally, they found that in interferon-activated locations near the sites of cardiac rupture, there was decreased expression of genes associated with fibroblast activation, which are necessary for robust cardiac wound healing and maintaining the integrity of the tissue.
With this data in hand, the authors suggest that mechanical strain in regions bordering cardiac injury leads to disruption of the nucleus and leakage of its contents into the cytoplasm of cardiomyocytes. These contents (such as nucleic acids) are then detected by innate defense machinery, triggering interferon signaling, and dampening fibroblast differentiation, leading to poor wound healing.
Graph Artificial Intelligence in Medicine [Johnson et al., Annual Review of Biomedical Data Science, August 2024]
A recent review published by the Zitnik Lab explores the growing role of graph-based artificial intelligence (AI) in medical applications. The review details how graph representation learning, particularly through graph neural networks (GNNs) and graph transformers, is revolutionizing the way complex clinical data is processed and analyzed.
The review highlights the natural fit of graph representations for modeling intricate relationships within clinical datasets. This approach allows for a more holistic view of patient health and medical knowledge, encompassing everything from patient records to imaging data. Graph-based models excel at transfer learning, enabling knowledge to be shared across different clinical tasks and patient populations—a crucial capability in healthcare where labeled data for specific conditions can be scarce.
One of the key strengths of GNNs is their ability to integrate diverse data types, including genomics, electronic health records, and medical imaging. This multimodal integration facilitates more comprehensive and personalized clinical predictions. Additionally, graph-based models offer inherent mechanisms for interpretability, which is essential for building trust among healthcare professionals and ensuring responsible AI deployment in clinical settings.
The authors emphasize how these models can effectively incorporate existing medical knowledge, such as biological pathways and medical ontologies. This integration enhances their predictive power and relevance to clinical practice. However, the review also addresses challenges in the field, including the need for improved scalability to handle large-scale biomedical datasets, strategies to address missing data in multimodal contexts, and the development of more sophisticated explainability techniques tailored to different healthcare stakeholders.
Looking to the future, the review suggests that graph AI models may evolve into foundation models for healthcare, capable of adapting to a wide range of clinical tasks with minimal fine-tuning. This development could significantly accelerate the adoption of AI in precision medicine and personalized healthcare.
What we listened to
Making our way through the Valence Labs and MILA machine learning for drug discovery summer school content. Two highlights:
What we liked on social
The impact of AI in drug discovery isn’t where you think it is @EricDai_BioE, @nc_frey
Things I learned talking to a new breed of scientific institutions @
ARPA-H project giving surgeons the power to see each cancer cell during resection @mattykirsh
Events
Build on cool datasets and models including ESM-3 at the Evolved hackathon
TechBio Transformers Berkeley meetup, Sept 27 @ 6pm
Jobs
Testing a new section out this week. Let us know what you think!
Senior Machine Learning Scientist @ Prescient Design/Genentech
Chief of Staff, Discovery Scientist, Lab Ops roles @ Arcadia Science
Fermentation Scientist @ Anthology
Bioengineering Intern @ Cradle Bio
Molecular Biology RA @ Neptune Bio