Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
A chunky one today… just like your July 4th celebrations… and the UK Labour party’s impending win…
What we read
Blogs
A Molecular Biologist’s Advice For Life [Bruce Booth, LifeSci VC, June 2024]
Reflecting on turning 50, Bruce Booth explores how science has been a central theme in his life. Inspired by campaigns like DuPont’s "Better Living Through Chemistry" and Pfizer's "Science Will Win," Bruce shares his advice by linking scientific concepts to life lessons:
Life’s Central Dogma: Prioritize self-care and personal growth to positively influence others.
Heterodimers: Combine diverse interests, like science and societal issues, to create impactful change.
Avoid Early Specialization: Stay broad and curious early in your career to build a strong foundation.
Embrace Change: Adapt to new opportunities and overcome personal inertia for long-term growth.
Pattern Recognition: Learn to recognize life’s patterns to make informed decisions and anticipate outcomes.
Social Immunity: Develop immunity against negative influences by following the No-Asshole-Rule (NAR).
Focus on Actions: Your actions and traits matter more than credentials or background.
Work-Life Symbiosis: Integrate personal passions with professional life for mutual benefit.
Friday Experiments: Set aside time for creative, non-linear thinking and experimentation.
Calculated Risks: Be bold in taking risks, but understand the potential downsides and benefits.
Celebrate Social Connections: Build deep professional and personal relationships for learning and growth.
Acknowledge Randomness: Recognize the role of luck and stay humble in your achievements.
B Cells as Drug Factories [Cormac Sheridann, Nature News, June 2024]
We now have early results from the world’s first clinical trial of B cell “biofactories”, which are essentially internal factories in the body that are engineered to produce a therapeutic protein continuously. These B cells are engineered to produce therapeutic proteins after differentiating into antibody-producing cells. It is a fascinating piece of scientific engineering that does not require immunosuppression or preconditioning, making it a safer way to treat disease and deliver genetic material. Immusoft and Be Biopharma are two companies paving the way. Immusoft’s Phase I trial for mucopolysaccharidosis type I involves B cells that are engineered to produce a-l-iduronidase and has shown promising early results. Be Bio is preparing an IND application for a hemophilia B treatment. While these advances bring optimism, there is still a long way to go for clinical acceptance and validating safety and efficacy for this new modality.
Papers
Bridge RNAs direct programmable recombination of target and donor DNA [Durrant et al, Nature 2024]
Back in january, scientists at the Arc Institute released a preprint on the discovery of bridgeRNAs - a new molecular system distinct from CRISPR that enables clean sequence-specific insertions, excisions and inversions to enable programmable genetic engineering. The team have now released two nature papers detailing the mechanism and structure of the system. The system is based on an IS110 recombinase and a bridge RNA. When the IS110 excises from the genome, the non-coding DNA ends join to form the bridge RNA that folds into two loops: one binding to IS110 and the other binding to the target DNA. Each loop of the bridge is independently programmable. The papers show cryo-EM structures of the recombinase-bridge RNA complex bound to DNA, sequentially stepping through the mechanism. The system promises cleaner editing and great predictability than the CRISPR-Cas9 system which and recently developed systems such as PASTE and PASSIGE require complicated components to be administered into the cell.
A high-density microfluidic bioreactor for the automated manufacturing of CAR T cells [Sin et al., Nature Biomedical Engineering, 2024]
CAR-T cell therapy manufacturing relies on either fed-batch and manual processes that lack a high degree of environmental control or is carried out on bioreactors that cannot be easily scaled out. Current CAR-T cell therapy manufacturing relies on centralized manufacturing. The long vein-to-vein turnaround times of 21-37 days have hindered patient access.
These centralized processes have many manual steps, which are prone to error, contamination and can lead to fluctuations in nutrients and waste elimination. Closed system bioreactors can run perfusion culture models with continuous medium circulation with improvements in cell growth and expansion. These systems, however, still require cleanroom facilities. This has led to increased efforts in a decentralized, automated point-of-care manufacturing of autologous cell therapies, which can decrease turnaround times to 8-13 days. Current decentralized systems, like the CliniMACS Prodigy and the Cocoon are still based on fed-batch cultures so they lack control of process parameters which are critical for T-cell expansion and differentiation.
In this paper, the authors show that primary T cells are activated, transducer and expanded in a 2ml automated closed system microfluidic bioreactor, commercialized as the Mobius Breeze, to produce viable anti-CD19 CAR-T cells with comparable activity to those produced in a gas-permeable well; meeting the minimum cell dose of Kymriah and exceeded the maximum cell dose of Yescarta.
The system incorporates integrated mixers, injectors and sensors for RT monitoring and control of “perfusion flow rate, optical density (OD), temperature, CO2, dissolved O2 and pH levels”. This innovation may improve the analysis of CAR-T cell phenotypes during culture and scale out of cell therapy manufacturing.
Brainwide silencing of prion protein by AAV-mediated delivery of an engineered compact epigenetic editor [Neumann et al., Science, June 2024]
A paper out in Science this week from the labs of Jonathan Weissman and Sonia Vallabh describes a novel approach to treating prion diseases using a compact epigenetic silencer called CHARM (Coupled Histone tail for Autoinhibition Release of Methyltransferase). Prion diseases are fatal neurodegenerative disorders caused by misfolded prion proteins, and currently lack effective treatments.
The method utilizes epigenetic modifications, specifically DNA methylation, to inhibit gene transcription without altering the underlying DNA sequence. CHARM consists of three key components:
A DNA-binding domain targeting the specific gene region
D3L protein machinery
A histone H3 tail
These components work together to guide the fusion protein to the target gene, activate DNA methylation, and silence the gene. The researchers delivered CHARM to mouse brains using an AAV vector. Treated mice exhibited increased DNA methylation at the prion protein promoter and significantly reduced prion RNA and protein levels.
Additionally, the team developed self-silencing CHARMs that deactivate after silencing their target. This feature limits CHARM expression temporally, potentially reducing antigenicity and off-target effects in nondividing neurons. A piece of target DNA was included in the AAV vector so that once CHARM also epigenetically silences itself in addition to the target gene.
Patient derived mini-colons enable long-term modeling of tumor-microenvironment complexity [Lorenzo-Martin et al., Nature Biotechnology, 2024]
Why it matters: Colorectal cancer (CRC) is the third most common cancer worldwide, accounting for approximately 10% of all cancer cases, and is the second leading cause of cancer-related deaths worldwide. In vitro models for CRC have remained lacking, largely due to their inability to properly recapitulate the tumor microenvironment. In this paper, the authors generate next-generation CRC organoids that enable more accurate assessment of interactions with cells from the cognate tumor microenvironment.Organoids have largely failed to model tumor microenvironments accurately, given there are several components that are hard to generate in vitro, including immune infiltration, vascularization, mechanobiology, and various components of the tumor stroma. Given these evolve over time, these also necessitate long-lived specimens, which are challenging to establish with current techniques. Here, the authors leverage an organ-on-a-chip device to enable scaffold-guided morganoid morphogenesis in microfluidic devices. This device has a micropatterned hydrogel to facilitate colonic structures such as crypts and lumens.
The authors seeded the chip with CRC cells (in a 1:99 ratio of cancer:healthy cells, recapitulating how cancer arises from a small number of cancer initiating cells). The healthy cells colonized the micropattern scaffold and formed an epithelium with a perfusable lumenal space, and cancer cells were subsequently integrated, leading to robust tumor growth. These mini-colons accurately mimicked several aspects of normal colon physiology, and were able to survive >1+ month, allowing the authors to follow the progression of emergent tumors. They then integrated autologous cancer-associated fibroblasts (CAFs) in the system, allowing them to migrate from an adjacent basal medium reservoir. CAFs successfully migrated into the tumor, and they were found to drive development of CRC invasive structures. Additionally, the authors were able to add tumor-infiltrating lymphocytes into another basal medium reservoir, which similarly infiltrated the hydrogel and migrated towards the CRC mini-colon and caused cancer regression over time.
In brief, the authors have demonstrated an ability to more accurately model the CRC tumor microenvironment over extended periods, using an organ-on-chip approach with a step-wise seeding of relevant TME cell types. However, an important caveat is that these take several weeks to generate, while clinical decision making in CRC can happen rapidly. Regardless, this represents an improvement over conventional organoids, and holds promise for assessing CRC dynamics.
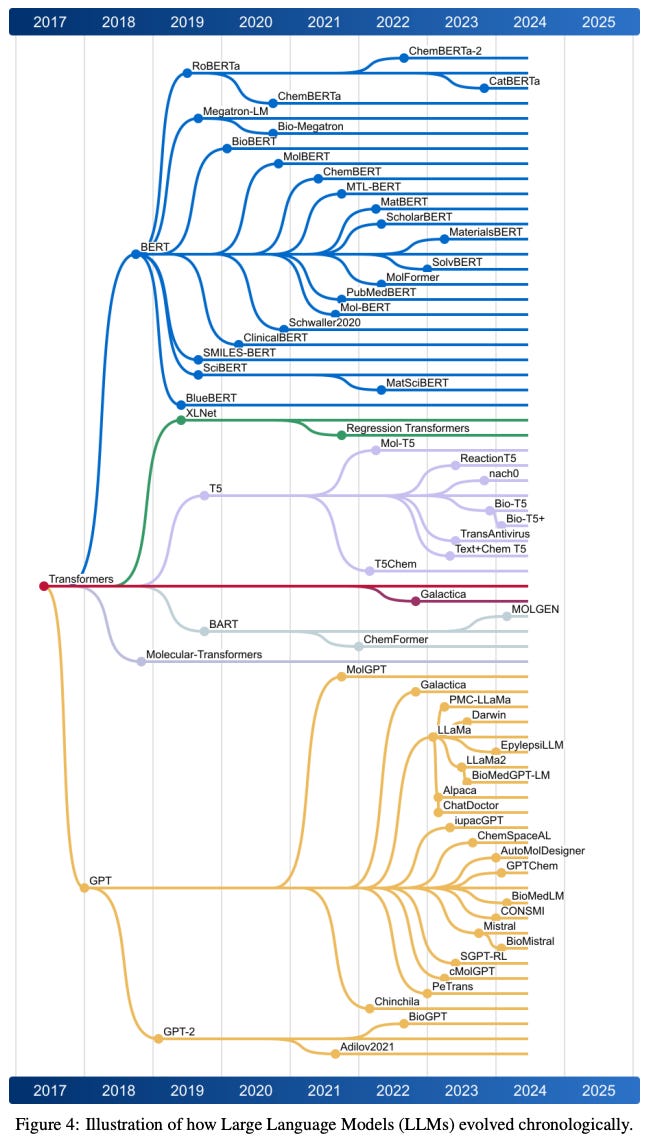
A Review of Large Language Models and Autonomous Agents in Chemistry [Ramos, Collison, White, arXiv, July 2024]
Why it matters: This review underscores the potential of LLMs and autonomous agents to revolutionize the field of chemistry. By automating complex tasks, these technologies can significantly accelerate scientific discovery and innovation. For instance, the ability to predict molecular properties and design new molecules quickly can lead to faster development of new drugs and materials. Moreover, the integration of these models into autonomous agents can streamline routine laboratory tasks, reduce human error, and free up researchers to focus on more creative and strategic aspects of their work. The paper also highlights the ongoing challenges in the field, such as the need for high-quality training data and the ethical considerations of AI-driven chemical research.This paper by the White lab explores the transformative role of large language models (LLMs) and autonomous agents in the field of chemistry. It highlights the significant advancements LLMs have brought to various domains within chemistry, such as accurate property prediction, molecule design, synthesis pathway optimization, and the acceleration of drug and material discovery. The authors delve into the integration of LLMs with chemistry-specific tools like synthesis planners and databases, leading to the development of autonomous agents that can perform complex chemical tasks with minimal human intervention.
The review covers the historical context and evolution of LLMs, current capabilities, and the specific challenges they face in the chemical domain. It emphasizes the importance of high-quality datasets for training these models and discusses various transformer architectures (encoder-only, decoder-only, and encoder-decoder) used in different chemistry applications. The paper also addresses the emergence of multi-agent systems and their potential to revolutionize experimental planning, literature review, chemical innovation, and cheminformatics tasks.
What we listened to
Notable Deals
Waypoint Bio raises $14.5M seed to bring spatial biology to CAR-T therapy - Two former MIT co-founders developed a groundbreaking technology that enables rapid, parallel testing of numerous cell therapies in mice. Their innovation uses detailed molecular maps to pinpoint where cell therapies encounter obstacles in attacking tumors, optimizing effective designs. The goal is to expedite cell therapy development by identifying successful designs and understanding their mechanisms. They aim to select a lead cell therapy by the end of 2025, with the recent funding round led by Hummingbird Ventures.
Granza Bio announces $7.14M seed - the Oxford / YC biotech is using a novel feature of the immune system that could enable better drug delivery
Beacon Therapeutics announces $170M Series B for a rare ocular gene therapy
ARPA-H enters $19M contract with Palantir for artificial intelligence, data software
What we liked on socials channels
Events
hsph.harvard.edu/pqg-conference/
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone