BioByte 032: AI and the overlooked stuff, base editing in mitochondria, ratio-tunable multi-gene expression, a brain-spine interface allows a man with tetraplegia to walk again
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
BIO is next week in Boston.. a few of us will also be out there—reach out and let’s meet up!
Biotech can’t stop, won’t stop:
Base editors just keep on coming. This time, we’re getting into the mitochondria and showing excellent efficiency, demonstrating applicability for therapy in mitochondrial genetic diseases.
Schizophrenia is a terrible disease with no real diagnostic test out there. Deep learning may enable the prediction and diagnosis of the disease by drawing out information from imaging modalities (EEG, fMRI, dMRI).
Synthetic biology will increasingly require more elegant and efficient systems for cell engineering. Scientists have developed a new method, SEMPER (stoichiometric expression of messenger polycistrons by eukaryotic ribosomes) to enable user-defined, tunable, multi-gene expression from single mRNAs.
A next-gen BSI (brain-spine interface) allows a patient with tetraplegia to walk independently and navigate complex terrain.
What we read
Blogs
AI and the Hard Stuff [Derek Lowe, May 2023]
A leading AI-driven drug discovery companies, BenevolentAI, recently underwent layoffs and restructuring in response to a PhIIa failure of their atopic dermatitis program. In a new essay, Derek Lowe used this failure as an opportunity to reiterate one of his primary criticisms of AI applications in drug discovery:
“There are no existing AI/ML systems that mitigate clinical failure risks due to target choice or toxicology.”
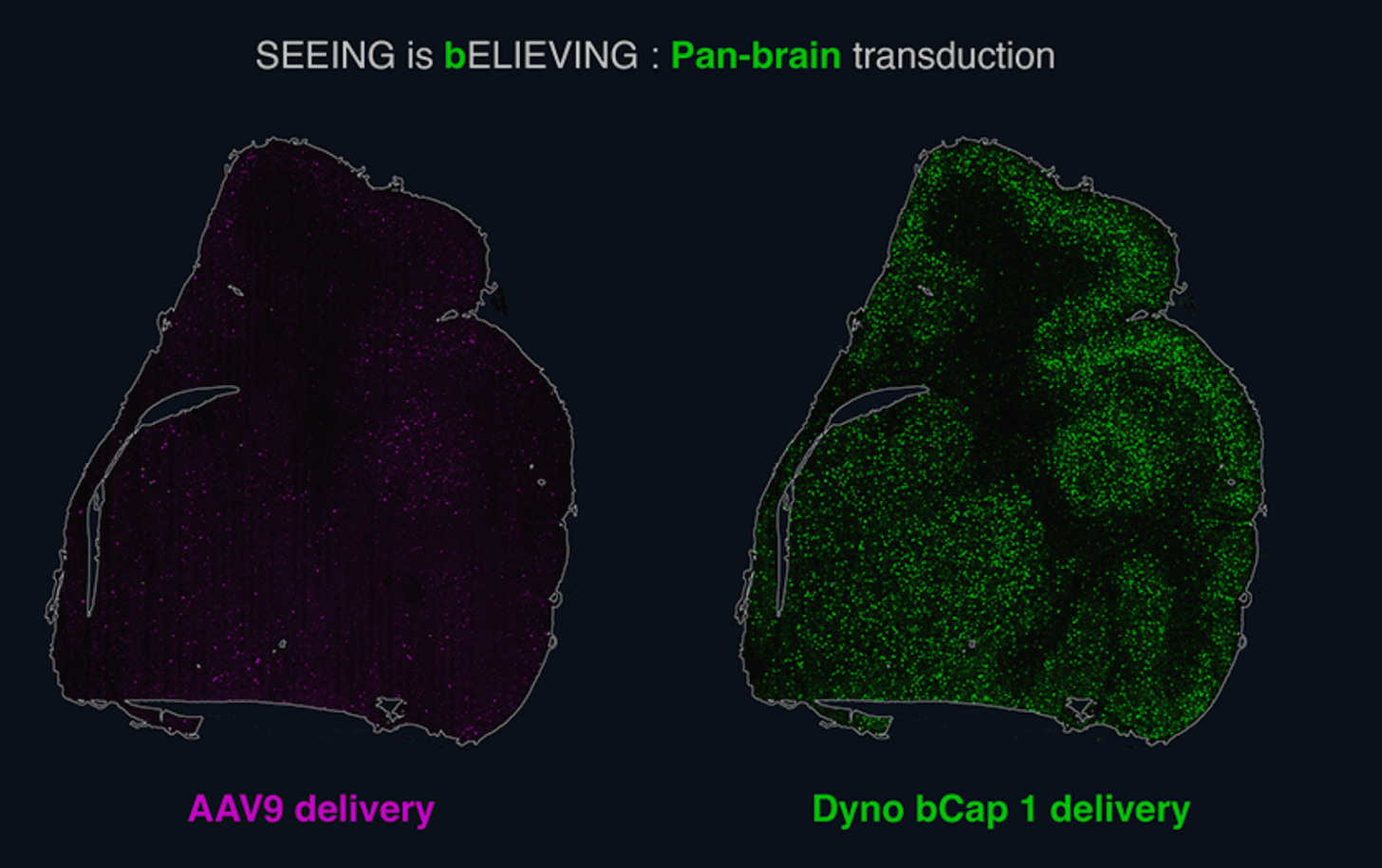
In general, Derek’s writing on the limitations of computational techniques in drug discovery (e.g., this essay on AlphaFold) are well-reasoned, and help ground an industry that, at times, is prone to exaggerated claims. In this case, however, the assessment that AI is only “being applied to lead compound generation, to patent-busting, to hit expansion” is incomplete, and overlooks recent work in early-stage biotech using computational techniques to derisk target biology or improve toxicity. For example, Dyno Therapeutics, an AI and wet-lab platform for designing cell-specific AAV vectors for gene therapy, recently presented NHP data at ASGCT on an AI-designed AAV capsid that can selectively target the CNS and detarget the liver (see figure below), improving biodistribution and addressing important toxicity concerns of systemically administered AAVs. Immunai is combining AI with single-cell multi-omics and experimental immunology to identify and validate novel targets, with the goal of decreasing the probability of clinical trial failure due to poor target choice.
It is misleading to imply that AI in drug discovery is mostly focused on patent-busting. There are many startups working hard to address the biggest bottlenecks of drug discovery: better characterizing target biology and improving toxicity profiles. To be sure, clinical evidence supporting the ability of AI to address these bottlenecks is early. AI is not a panacea; it’s just a tool. A primary goal of this newsletter is evaluating and communicating the emerging evidence around the efficacy of this tool, both good and bad.
The Disparity in UI/UX between Biotech & Tech [Vega Shah, The Aliquot, 2023]
Any of you that have worked in a lab (or clinical setting) will commiserate: software feels like it is light years behind what we use on a daily basis coming out from the tech world. Why do we continually have to deal with poor UI/UX? Last year, Vega asked the Twitterverse why biotech UI/UX lags behind those in big tech; she breaks down responses from industry leaders.
Biotech companies tend to prioritize function over form; this culture likely stems from its roots in academic research.
Even software-oriented domains of biotech (such as bioinformatics) haven’t prioritized aesthetics; students undergo their training and become used to cumbersome software. Thus, the bar for aesthetics is truly on the floor.
The cost of creating, disseminating, and garnering adoption of new software is too great. Again, if it doesn’t offer a significant advantage in daily lab workflows, people will rarely bother. Additionally, migrating from one software to another comes with its own challenges: regulatory compliance, ensuring the robust migration of precious data, and more.
Biotech companies have difficulty drawing, training, and retaining UI/UX talent. With significant competition from big tech companies (which are often better capitalized and can pay better, and also have a significant internal focus on aesthetics/usability), it remains difficult to draw talent that will continue to push forward UI/UX.
Techbio is realizing the dream of big data [Amulya Garimella, May 2023]
In this essay, Amulya draws parallels between the field of data science in tech and biotech. She argues that bio companies can “finally fulfill the promise of Big Data: finding hidden patterns in the vast, complex mess” and a lot of this is because in amphibious tech x bio companies, biologists and computationalists are side by side innovating together. An assay is created, the results are analyzed, and the data is fed back into the system. There is less waiting around and losing unstructured data. While we acknowledge that this generalization is grossly oversimplified, it represents a key culture shift from biotech of the past to the types of companies we are currently seeing push major innovation—biologists and computationalists must work together in tandem. It’s not one or the other, it’s not before or after, it’s together.
Benchtop DNA printers are coming soon—and biosecurity experts are worried [Service, Science, May 2023]
Advances in DNA synthesis technology are enabling biologists to conveniently print all the DNA they need using benchtop machines, mitigating the need to obtain DNA sequences online from the ~100 companies that provide DNA printing and shipping services. However, this development poses new risks as it bypasses the existing screening processes employed by synthetic biology companies to detect potential bioterrorists.
A recent report released by the Nuclear Threat Initiative (NTI), a Washington, D.C.-based think tank, emphasizes the need for stronger safeguards in order to prevent misuse of this technology by malicious actors or accidental catastrophic incidents. The report recommends that governments, industry, and the scientific community revamp screening procedures and implement stricter regulations. While the technology offers benefits for genetic research, concerns arise due to the potential misuse and the ease of splicing large pathogen genomes. The report suggests customer vetting, pre-synthesis screening software, updated guidelines, and mandatory requirements as potential solutions. The urgency to address these concerns is emphasized, with experts highlighting the need for swift action to ensure safety and security in the face of advancing benchtop synthesis technology.
“Governments, industry, and the broader scientific community need to put stronger safeguards in place to ensure this technology is not exploited by malicious actors and that it doesn’t lead to a catastrophic accident.”
-Jaime Yassif, vice president for global biological policy and programs at the Nuclear Threat Initiative
Check out the long-form report from NTI on Benchtop DNA Synthesis Devices: Capabilities, Biosecurity Implications, and Governance here.
Is it Time to Cancel Menopause [Grace Rubenstein, proto.Life, Jun 2022]
With all the recent buzz on ovarian aging and epigenetic reprogramming, we’re digging up this piece by Grace Rubenstein that first appeared last year. Grace takes a deep dive into technologies centered around ovarian aging, notably staving off menopause to extend fertility and increase the quality of life of women. She sheds light on an idea that’s becoming increasingly popular—using the ovary as a time machine to study broader longevity pathways given that the ovaries age 2-3 times faster than the rest of the human body.
In practice, Rubenstein asks Jennifer Garrison, professor at the Buck Institute, on where insights can come from:
Trying biochemical interventions designed for anti-aging on the ovaries (gives rise to experiments interrogating things like rapamycin and NAD+ on ovarian function)
Examining environmental contaminants that may affect ovarian function and egg quality (pesticides, hormone disruptors, plastics, etc.)
Finding cellular targets related to inflammation, metabolic function, etc. (for example, examining senescent cells)
Developing biomarkers to chart fertility age
It’s unclear which approaches will prove fruitful given the fundamental problem underlying fertility—we don’t know enough about how the natural processes happen in our bodies. The signals and physiological changes are still largely mysteries. Some of these suggested experiments shed light on the natural processes so that we can better engineer them. Does staving off menopause mean arresting eggs to trick the body into thinking fertility status has been extended? Does it mean inserting artificial new oocytes into the ovaries? Does it look like an entirely different class of cell therapy or hormone therapy? The possibilities are endless as are the questions stopping them. We’re excited to keep a close eye on this field.
Academic papers
Strand-selective base editing of human mitochondrial DNA using mitoBEs [Yi et al., Nature Biotechnology, 2023]
[Why it matters] Several diseases are driven by point mutations in the mitochondria; however, it has proven difficult to deliver gene editing machinery to the organelle. In this paper, the authors deliver mitochondrial base editors (encoded in circular RNA), and demonstrate efficient editing with applicability for treating mitochondrial genetic diseases. In line with our 2023 prediction, this week we have a new exciting gene editing system to cover. Mitochondrial DNA base editors (or mitoBEs) combine a TALEN and a deaminase for precise base editing in mitochondrial DNA.
95% of disease-associated mtDNA mutations are point mutations which could be corrected using gene editing approaches. However, the delivery of CRISPR guide RNAs into the mitochondria is difficult. In order to overcome these challenges, the team developed a programmable TALEN-based base editor by fusing it to either a ssDNA-specific adenine deaminase or a cytosine deaminase. They achieved A-to-G or C-to-T base editing with up to 77% efficiency. To deliver the base editors they used circular RNA (circRNA) to encode them. The team demonstrated strand-biased editing in various human cell types, indicating compatibility with the circRNA delivery route.
A Survey on the Role of Artificial Intelligence in the Prediction and Diagnosis of Schizophrenia [Ramesh et al., arXiv, 2023]
[Why it matters] Schizophrenia is a devastating mental disorder with no real diagnostic tests. With an extended prodromal phase, there remains a significant need to identify patients at their earliest onset of symptoms to help treat and mitigate some of the downstream sequelae. This review succinctly summarizes advances made the in the deep learning field for diagnosing schizophrenia; although early, there is promise for using compute to more quantitatively assess patients for their risk of developing schizophrenia. In this survey, the authors review studies that use deep learning to detect and predict schizophrenia using EEG, fMRI and dMRI. Schizophrenia affects 1 in 300 people, with an estimated prevalence of 24 million people worldwide. The disorder cannot be diagnosed using a clinical test and is rather done through the monitoring of changes in the patient’s behavior. Whilst diagnosis is challenging due to the lack of quantitative tools, behavior-based diagnosis and adequate treatment can have a profound effect on the patients quality of life. This 10-paper survey showed that the use of deep learning (mainly RNNs, CNNs and SVMs) applied to EEG/MRI can lead to successful predictions of more than 80%.
Stoichiometric expression of messenger polycistrons by eukaryotic ribosomes (SEMPER) for compact, ratio-tunable multi-gene expression from single mRNAs [Duan et al., bioRxiv, 2023]
[Why it matters] The increasing complexity of mammalian synthetic biology – whether the production of synthetic circuits, protein assemblies, or more – necessitates the ability to produce multiple proteins with controllable stoichiometries with simple and elegant engineering. Duan et al., develop a method, SEMPER, that allows for the expression of multiple ORFs from a single transcript, with tunable ratios. This technology represents a step-change in the ease of engineering tunable expression of transcripts in simple eukaryotic constructs.Current approaches to producing multiple transcripts at required stoichiometries are unwieldy, such as using large constructs with promoter engineering or gene copy number titration. These are large, limiting their ability to be delivered to cells. Here, the authors develop SEMPER, a method taking advantage of leaky ribosome scanning (LRS), enabling compact, tunable, polycistronic expression in mammalian cells using plasmid and mRNA-based expression. Introduction of a short ORF (opening reading frame) just upstream of a gene of interest (GOI) has been shown to push mammalian cells to using LRS and alternate translation initiation sites (TISs) to shunt ribosomes away from a GOI. By replacing non-protein-coding uORFs with longer coding sequences, tuning the translation initiation strength of each ORF, and chaining them together, this approach enables polycystronic expression of multiple proteins with user-defined translation rates. The authors additionally demonstrate the ability to encode multi-gene constructs and can be applied directly to mRNA-based circuits using IVT.
An Analysis of Successful Hit-to-Clinical Candidate Pairs [Brown, Journal of Medicinal Chemistry, 2023]
[Why it matters] A recent study analyzed 156 clinical candidates published in the Journal of Medicinal Chemistry between 2018 and 2021 to identify common strategies used to generate lead compounds for drug development. The analysis found that the most frequent lead generation strategies resulting in clinical candidates were based on known compounds (59%) and random screening approaches (21%). Other approaches included directed screening, fragment screening, DNA-encoded library screening (DEL), and virtual screening.The study also examined the similarity between the clinical candidates and their original hits using Tanimoto-MCS, revealing that most clinical candidates were structurally different from their hits but shared a key pharmacophore, which is the spatial arrangement of atoms or functional groups in a molecule that is responsible for its biological activity and interaction with a target receptor (and therefore desired pharmacological effect). Additionally, the study examined the incorporation of oxygen, nitrogen, fluorine, chlorine, and sulfur in clinical candidates. The three most similar and least similar hit-to-clinical pairs from random screening were further analyzed to gain insights into the changes that lead to successful clinical candidates.
The results from the study suggest that starting from existing clinical candidates or published scaffolds can guide the optimization process and highlight the importance of early candidates in understanding pharmacokinetics/pharmacodynamics (PK/PD) and efficacy drivers. While random screening still contributes a significant number of starting points, most clinical candidates require further optimization. During optimization, the molecular weight (MW) tends to increase, but lipophilicity remains relatively unchanged. Fluorine incorporation is the most common, with limited use of heavier halogen atoms or sulfur. Most clinical candidates adhere to the Rule of Five (Ro5), with MW often being the only violation.
As a fun, quick refresher, recall that the Rule of Five (aka the Lipinkski rule) in medicinal chemistry helps to predict if a biologically active molecule is likely to have the chemical and physical properties to be orally bioavailable and therefore suitable for a drug. The rule utilizes pharmacokinetic drug properties such as absorption, distribution, metabolism and excretion on specific physicochemical properties such as:
No more than 5 hydrogen bond donors
No more than 10 hydrogen bond acceptors
Molecular mass less than 500 Da
Partition coefficient not greater than 5
According to the rule, an orally active drug can have no more than one violation of these conditions. Now, you might by wondering where the name of the rule came from. It turns out, all the conditions have multiples of five as the determinant conditions!
Walking naturally after spinal cord injury using a brain–spine interface [Lorach et al., Nature, May 2023]
[Why it matters] In a first for the BCI field, a top neurorehabilitation and prosthetic lab develops a closed-loop system comprising an ECoG array in sensorimotor cortex and epidural spinal cord stimulator, enabling a patient with tetraplegia to independently navigate complex, natural environments. In the latest paper from the NeuroX Institute in Switzerland, researchers engineered an implantable recording and stimulation system that enabled an individual with chronic tetraplegia to stand and walk in natural settings. The system consisted of 2 parts:
Two cortical electrocorticographic (ECoG, or a strip of EEG electrodes) arrays positioned epidurally over the sensorimotor cortex, with a simple ML algorithm to decode motor intentions.
A spinal cord stimulator that translates these motor signals into patterns of epidural stimulation over the dorsal root entry zones of the spinal cord.
To determine where to optimally implant the ECoG arrays to successfully decode motor intentions, anatomical and functional imaging data was used. The system was calibrated in just a few minutes by recording ECoG signals while asking the participant to attempt movement while seated. The participant was able to walk independently, including navigating complex terrain like stairs. Importantly, the system and cortical signals were stable over time, and the participant successfully used the device with no changes in stimulation thresholds for over one year of use. Check out this amazing video of the participant walking using the BCI system.
What we listened to
Notable Deals
Modulo launches with $8M to reprogram the neuroimmune system
Larkspur Biosciences kicks off with $30M+, hunting for new oncology immunotherapies
BenevolentAI CFO resigns as company lays off staff, restructures
ElevateBio raises $401M Series D in the largest biotech funding round of 2023
In case you missed it
New prediction categories in CASP15 [Kryshtafovych et al., Authorea Preprints, 2023]
What we liked on Twitter
Events
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone[subscribe button]
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone













"Biotech companies have difficulty drawing, training, and retaining UI/UX talent."
I can't speak for the current situation but several years ago, biotech and healthtech companies showed little interest. IIRC on Linkedin there was just one UX/UI position at Ginkgo.